MPDB Presentation
- 1. MPDB - Integrated system for storage and analysis of metabolomic data Design and implementation of the data acquisition and analysis pipeline Alexander Raskind, SFRES MTU
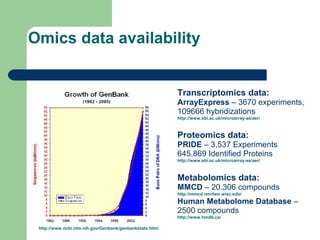
- 2. Omics data availability http://www.ncbi.nlm.nih.gov/Genbank/genbankstats.html Transcriptomics data: ArrayExpress â 3670 experiments, 109666 hybridizations http://www.ebi.ac.uk/microarray-as/aer/ Proteomics data: PRIDE â 3,537 Experiments 645,869 Identified Proteins http://www.ebi.ac.uk/microarray-as/aer/ Metabolomics data: MMCD â 20.306 compounds http://mmcd.nmrfam.wisc.edu/ Human Metabolome Database â 2500 compounds http://www.hmdb.ca/
- 3. Shifting research paradigm genome.uiowa.edu http://www.shimadzu.com Targeted analysis High-throughput analysis
- 4. Populus as model system âĒ Wide ecological range âĒ Small genome relative to other trees âĒ Relatively easy transformation and cloning âĒ Belongs to Salicaceae â Willow family, produces large amount of phenolic compounds that may influence carbon sequestration
- 5. Project rationale âĒ Affordable equipment generates limited amount of metabolomic data with modest quality âĒ Proper information storage and maximal extraction of useful information are essential âĒ Free open source laboratory information system tailored to metabolomics workflow would benefit to a large scientific community
- 6. System requirements âĒ Easy access to large arrays of analytical results and biological metadata âĒ Tools for data analysis âĒ Addition of analysis modules âĒ Accommodation of other types of analytical data âĒ USER FRIENDLY
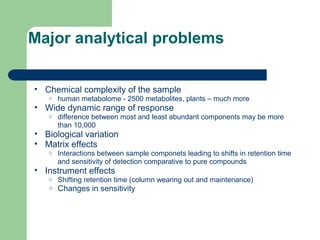
- 8. Major analytical problems âĒ Chemical complexity of the sample o human metabolome - 2500 metabolites, plants â much more âĒ Wide dynamic range of response o difference between most and least abundant components may be more than 10,000 âĒ Biological variation âĒ Matrix effects o Interactions between sample componets leading to shifts in retention time and sensitivity of detection comparative to pure compounds âĒ Instrument effects o Shifting retention time (column wearing out and maintenance) o Changes in sensitivity
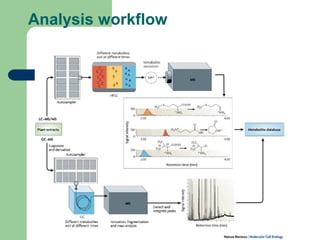
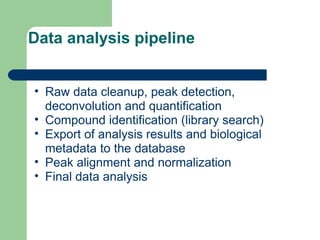
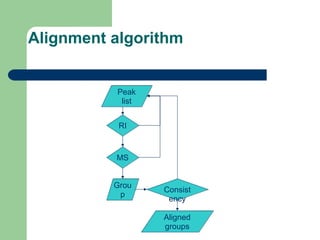
- 9. Data analysis pipeline âĒ Raw data cleanup, peak detection, deconvolution and quantification âĒ Compound identification (library search) âĒ Export of analysis results and biological metadata to the database âĒ Peak alignment and normalization âĒ Final data analysis
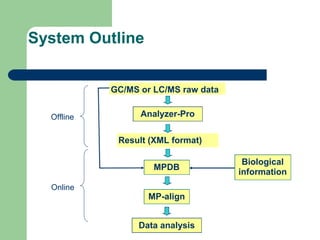
- 10. System Outline Analyzer-Pro Result (XML format) MP-align GC/MS or LC/MS raw data MPDB Offline Online Data analysis Biological information
- 11. Compound identification âĒ NIST 2002 database for GCMS (MS only, ~140,000 entries) âĒ In-house database of essential metabolites (MS and retention time, ~200 entries)
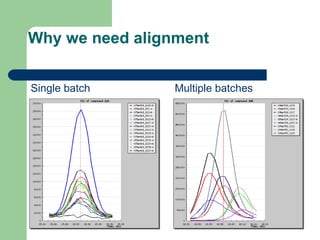
- 12. Why we need alignment Single batch Multiple batches
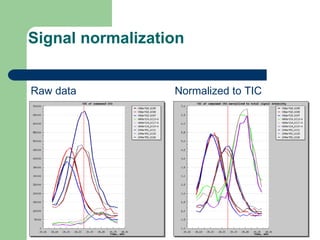
- 15. Signal normalization Raw data Normalized to TIC
- 16. User interface - tasks âĒ Data entry âĒ New analysis âĒ Review analysis âĒ Quality control âĒ Help
- 18. Sample groups review and annotation
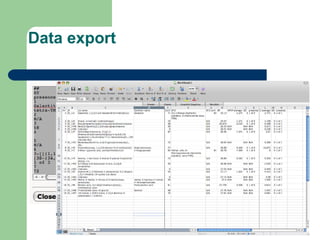
- 20. Data export
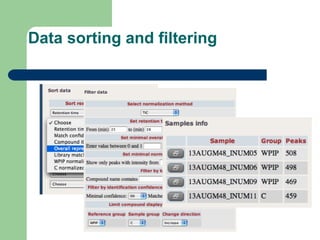
- 21. Data sorting and filtering
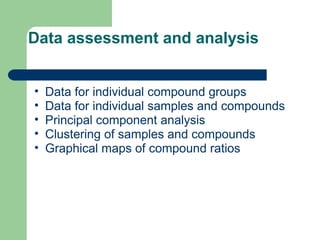
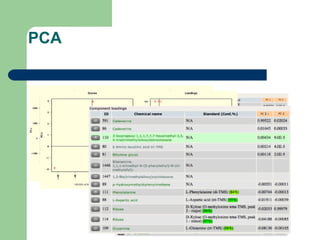
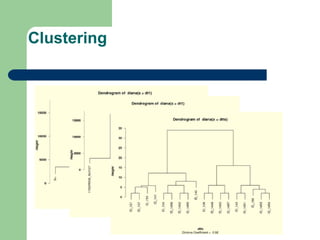
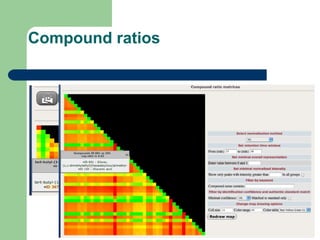
- 22. Data assessment and analysis âĒ Data for individual compound groups âĒ Data for individual samples and compounds âĒ Principal component analysis âĒ Clustering of samples and compounds âĒ Graphical maps of compound ratios
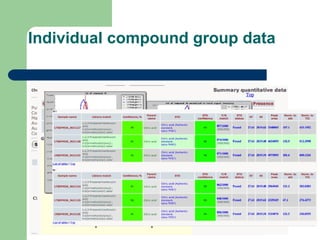
- 23. Individual compound group data
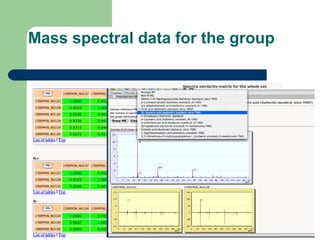
- 24. Mass spectral data for the group
- 25. Individual sample and peak details
- 26. PCA
- 27. Clustering
- 28. Compound ratios
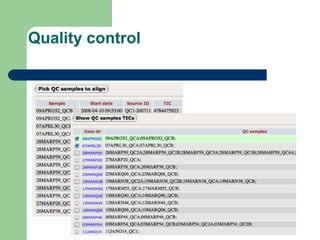
- 29. Quality control
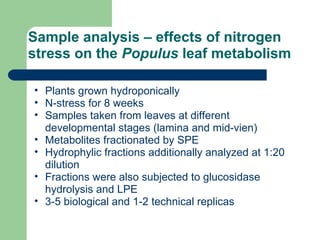
- 30. Sample analysis â effects of nitrogen stress on the Populus leaf metabolism âĒ Plants grown hydroponically âĒ N-stress for 8 weeks âĒ Samples taken from leaves at different developmental stages (lamina and mid-vien) âĒ Metabolites fractionated by SPE âĒ Hydrophylic fractions additionally analyzed at 1:20 dilution âĒ Fractions were also subjected to glucosidase hydrolysis and LPE âĒ 3-5 biological and 1-2 technical replicas
- 31. Leaf hydrophilic fraction âĒ Up-regulated by N-stress: o Galacturonic acid (X7), D-Arabinonate, o Turanose, Syringin o Ribose(?), methyl-Galactoside, 3-Hydroxy-3- methylglutaric acid (HMGA), D-(-)-3- Phosphoglyceric acid
- 32. Leaf hydrophilic fraction âĒ Down-regulated by N-stress: o Most of free aminoacids and polyamines below detection level or strongly reduced. Also some sugars and polyols, but not clearly identified) o Small organic acids (fumaric, succinic, threonic, citric, malic, oxaloacetic) o Sugar phosphates (glucose, fructose) o Xylose, melibiose, cellobiose
- 33. Acknowledgements âĒ Prof. Scott Harding âĒ Prof. Chung-Jui Tsai âĒ Dr. Changyu Hu âĒ Prof. Meir Edelman (WIS)