2018 ABRF Tools for improving rigor and reproducibility in bioinformatics
- 1. Stephen D. Turner, Ph.D. Bioinformatics Core Director University of Virginia School of Medicine bioinformatics.virginia.edu @strnr Tools for Improving Rigor & Reproducibility in Bioinformatics ║▌║▌▀Żs: bit.ly/madssci2018repro
- 2. We Are in the Middle of a New Movement in Genomics ŌĆó Genomics/bioinformatics advancing at grueling pace - New questions - New study designs - New technologies, new [’¼üll-in-the-blank]-seq ŌĆó New movements have: - Leaders / method developers / early adopters - First followers - Everybody else ŌĆó New technology leads to more reproducibility issues 2 CORES!
- 3. Reproducibility is hard! ŌĆó Genomics data is too large and high dimensional to easily inspect or visualize. ŌĆó Work’¼éows involve multiple steps and it's hard to inspect every step. ŌĆó Unlike in the wet lab, we don't always know what to expect of our genomics data analysis. ŌĆó It can be hard to distinguish good from bad results. 3
- 4. 4
- 5. Reproducibility:ŌĆ© What's in it for you? ŌĆó Your future self will thank you - Re-running analysis with different parameters - Re-running analysis with new data - Documentation ŌĆó Faster/cheaper - Modular work’¼éows - Reusable code chunks ŌĆó Makes collaboration with others easier 5 "Robust research is about doing small things that stack the deck in your favor to prevent mistakes." ŌĆōVince Buffalo, author of Bioinformatics Data Skills (2015).
- 6. Obstacles to Reproducibility 1. Bioinformatics software 2. Pipeline / work’¼éow management 3. Documentation 4. Data / code sharing 6 A non-comprehensive list of
- 8. Bioinformatics Software ŌĆó Bioinformatics software implements complex algorithms. - Dozens of parameters, endless permutations - Defaults not always optimal ŌĆó Perception: 8 ACACTCGCATCCGCACATCGCACTA GGTCAGCATACGCCGACTCCGACCG GCGCTATCGCCAGCGGAAATCGCAA
- 9. Bioinformatics Software ŌĆó Bioinformatics software implements complex algorithms. - Dozens of parameters, endless permutations - Defaults not always optimal ŌĆó Reality: Software is written by smart people, but: - Not software engineers - Not using good practice (version control, modularization, commentary, testing) - Unable to offer long-term ŌĆ© maintenance / support - Focus on graduating / ŌĆ© publishing, not support - Not always easily available 9
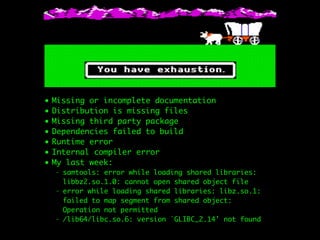
- 10. ŌĆó Missing or incomplete documentation ŌĆó Distribution is missing files ŌĆó Missing third party package ŌĆó Dependencies failed to build ŌĆó Runtime error ŌĆó Internal compiler error ŌĆó My last week: - samtools: error while loading shared libraries: libbz2.so.1.0: cannot open shared object file - error while loading shared libraries: libz.so.1: failed to map segment from shared object: Operation not permitted - /lib64/libc.so.6: version `GLIBC_2.14' not found
- 12. Package managers 12 Mac OS WindowsLinux apt-get yum homebrew macports ????? ????? Cross-platform
- 13. Conda ŌĆó Cross-platform package manager: Win, Mac, Linux ŌĆó Language agnostic (can be used to install C/C++, Fortran, Go, R, Python, Perl, Java, etc.). ŌĆó User-installable ŌĆō no admin/root privileges needed. ŌĆó Describes packages with a recipe de’¼üning dependencies and a build script that installs. ŌĆó Channels: conda provides many common packages by default. Additional channels add more. ŌĆó Isolated environments - Versions and tools can be managed per-project - No con’¼éicts or version incompatibility - Environments can be shared via simple text ’¼üles 13
- 14. Conda: Main commands ŌĆó conda create -n <environment> ŌĆó source activate <environment> ŌĆó conda search <package> ŌĆó conda install <package> ŌĆó conda upgrade <package> ŌĆó conda uninstall <package> 14
- 15. Conda: example ŌĆó Create a new environment named madssci:ŌĆ© conda create -n madssci ŌĆó Activate that environmentŌĆ© source activate madssci ŌĆó Install some packagesŌĆ© conda install blast bioconductor-flowcore ŌĆó Install a particular versionŌĆ© conda install samtools=0.1.19 15
- 16. Bioconda ŌĆó bioconda.github.io ŌĆó Bioconda is a channel for the conda package manager ŌĆó Repository for more than 3,000 bioinformatics packages ready to use with conda install ŌĆó >250 contributors have added/updated recipes ŌĆó Preprint: Gr├╝ning, Bj├Črn, et al. "Bioconda: A sustainable and comprehensive software distribution for the life sciences."┬ĀbioRxiv┬Ā(2017): 207092.ŌĆ© https://www.biorxiv.org/content/early/2017/10/27/207092 ŌĆó See also: "Nature TechBlog: Bioconda Promises to Ease Bioinformatics Software Installation Woes" ŌĆ© http://blogs.nature.com/naturejobs/2017/11/03/techblog-bioconda- promises-to-ease-bioinformatics-software-installation-woes/ 16
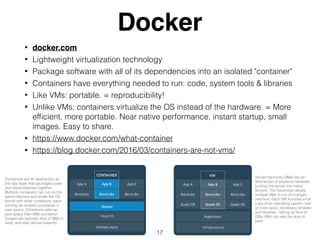
- 17. Docker ŌĆó docker.com ŌĆó Lightweight virtualization technology ŌĆó Package software with all of its dependencies into an isolated "container" ŌĆó Containers have everything needed to run: code, system tools & libraries ŌĆó Like VMs: portable. = reproducibility! ŌĆó Unlike VMs: containers virtualize the OS instead of the hardware. = More ef’¼ücient, more portable. Near native performance, instant startup, small images. Easy to share. ŌĆó https://www.docker.com/what-container ŌĆó https://blog.docker.com/2016/03/containers-are-not-vms/ 17 Containers are an abstraction at the app layer that packages code and dependencies together. Multiple containers can run on the same machine and share the OS kernel with other containers, each running as isolated processes in user space. Containers take up less space than VMs (container images are typically tens of MBs in size), and start almost instantly. Virtual machines (VMs) are an abstraction of physical hardware turning one server into many servers. The hypervisor allows multiple VMs to run on a single machine. Each VM includes a full copy of an operating system, one or more apps, necessary binaries and libraries - taking up tens of GBs. VMs can also be slow to boot.
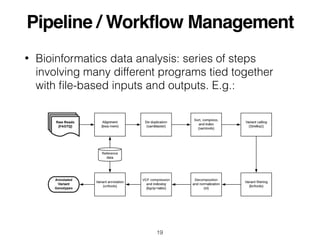
- 19. Pipeline / Workflow Management ŌĆó Bioinformatics data analysis: series of steps involving many different programs tied together with ’¼üle-based inputs and outputs. E.g.: 19
- 20. Pipeline / Workflow Management ŌĆó Simple solution: simple (bash) script - List of commands - Pros: quick, easy, portable, universal - Cons: not scalable, no re-entry / partial execution, assumes dependency availability, dif’¼ücult / no parallelization ŌĆó Work’¼éow management systems - Make (installed on most systems) - Snakemake - Next’¼éow - Galaxy - Many more: github.com/pditommaso/awesome-pipeline 20
- 21. Nextflow ŌĆó next’¼éow.io ŌĆó Di Tommaso, Paolo, et al. "Next’¼éow enables reproducible computational work’¼éows." Nature biotechnology 35.4 (2017): 316-319. ŌĆó Features: - Free - Actively developed - Supports docker containers - Easy parallelization, implicitly de’¼üned - Continuous checkpointing & resumed execution - Easily portable across architectures (SGE, LSF, SLURM, PBS, Amazon AWS, ...). 21
- 22. Beware of Pipelineitis ŌĆó ŌĆ£PipelinesŌĆØ can kill your creativity and force you to think too rigidly. ŌĆó DonŌĆÖt ŌĆ£pipelineŌĆØ too early, if at all. ŌĆó Does it even need to be pipeline-i’¼üed? ŌĆó WhoŌĆÖs running it? - You, once: donŌĆÖt pipeline-ify. Document, move along. - You, 2-5 times: documented script? - You, 10+ times: consider pipeline-ifying. - Others: create sharable pipeline ŌĆó See: Loman & Watson. "So you want to be a computational biologist?" Nat Biotechnol 31 (2013): 996-998. 22
- 23. Documentation 23
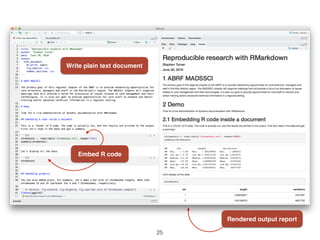
- 24. Dynamic Documentation: RMarkdown ŌĆó R: widely used for data science & bioinformatics ŌĆó Markdown: a simple markup language that allows you to render structured/formatted documents from plain text. ŌĆó RMarkdown: embeds R code in a Markdown document. - Write documents that execute embedded code and integrates results into the ’¼ünal report. - Allows you to keep code and documentation together. - Easily re-render the document, re-running analysis and re-incorporating results on the ’¼éy. - Many output formats: PDF, DOCX, HTML, EPUB, ... 24
- 25. 25 Write plain text document Embed R code Rendered output report
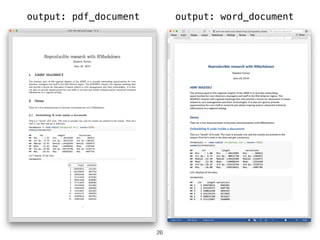
- 26. 26 output: pdf_document output: word_document
- 27. Jupyter notebooks ŌĆó jupyter.org ŌĆó Jupyter: open source project to develop software, standards, services across many languages ŌĆó Jupyter notebook: free application to create documents containing live code, visualizations, narrative text. - Supports >40 programming languages - Easily shared - Interactive output - Multi-user versions for companies, classrooms, labs 27
- 28. Data / code sharing 28
- 29. Sharing Code ŌĆó State of the art early-2000's - "Data/code available upon request" - "Code available on <lab website>" - None of the above ŌĆó Schultheiss, Sebastian J., et al. "Persistence and availability of web services in computational biology."┬ĀPloS one┬Ā6.9 (2011): e24914. - Surveyed ~1000 web services published in NAR 2003-2009 - ~30% unavailable - ~80% developed by students / non-permanent researcher ŌĆó Russell, Pamela H., et al. "A large-scale analysis of bioinformatics code on GitHub."┬ĀbioRxiv┬Ā(2018): 321919. ŌĆó github.com is becoming the de facto standard for archiving and sharing code 29
- 30. Sharing any research output 30 - ’¼ügshare.com - Free - Upload any ’¼üle format - Get a DOI - 5 GB max ’¼üle size - 20GB private space - Unlimited public space - Launched 2012 - Hosted on S3, multiple redundant copies - SLA: 10 yr persistence - zenodo.org - Free - Upload any ’¼üle format - Get a DOI - 50 GB per record - Higher quota by request - Unlimited records - Launched 2013 - Hosted at CERN (est 1954), with de’¼üned program of Ōēź20 years - about.zenodo.org/policies/ - about.zenodo.org/principles/ - osf.io - Free - Upload any ’¼üle format - Get a DOI - 5 GB per ’¼üle - Connect to any external storage provider - Launched 2013 - Preservation fund guaranteeing 50+ years of persistent availability - osf.io/faq
- 32. Other Resources 32 Wilson, et al. "Good enough practices in scientific computing." PLoS computational biology 13.6 (2017): e1005510. Wilson, et al. "Best practices for scientific computing." PLoS biology 12.1 (2014): e1001745. https://doi.org/10.1371/journal.pbio.1001745 https://doi.org/10.1371/journal.pcbi.1005510
- 33. Other Resources ŌĆó 2017: Ten simple rules for making research software more robust: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005412 ŌĆó 2017: Ten simple rules for responsible big data research: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005399 ŌĆó 2017: Ten Simple Rules to Enable Multi-site Collaborations through Data Sharing: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005278 ŌĆó 2016: Ten Simple Rules for Digital Data Storage: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005097 ŌĆó 2016: Ten Simple Rules for Effective Statistical Practice: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004961 ŌĆó 2015: Ten Simple Rules for Creating a Good Data Management Plan: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004525 ŌĆó 2015: Ten Simple Rules for ExperimentsŌĆÖ Provenance: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004384 ŌĆó 2015: Ten Simple Rules for a Computational BiologistŌĆÖs Laboratory Notebook: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004385 ŌĆó 2015: Ten Simple Rules for Reducing Overoptimistic Reporting in Methodological Computational Research: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004191 ŌĆó 2014: Ten Simple Rules for the Care and Feeding of Scienti’¼üc Data: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003542 ŌĆó 2014: Ten Simple Rules for Effective Computational Research: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003506 ŌĆó 2013: Ten Simple Rules for Reproducible Computational Research: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003285 ŌĆó 2012: Ten Simple Rules for the Open Development of Scienti’¼üc Software: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002802 ŌĆó 2014: Ten Simple Rules for Writing a PLOS Ten Simple Rules Article: ŌĆ© http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003858 33 collections.plos.org/ ten-simple-rules
- 34. Other Resources ŌĆó Baker, Monya. ŌĆ£1,500 Scientists Lift the Lid on Reproducibility.ŌĆØ Nature News, vol. 533, no. 7604, May 2016, p. 452. www.nature.com, doi:10.1038/533452a. ŌĆó Gr├╝ning, Bj├Črn, et al. ŌĆ£Practical Computational Reproducibility in the Life Sciences.ŌĆØ BioRxiv, Oct. 2017, p. 200683. www.biorxiv.org, doi:10.1101/200683. ŌĆó Leek, Jeff. "A Few Things That Would Reduce Stress around Reproducibility/ Replicability in Science." Simply Statistics, November 2017: https://simplystatistics.org/ 2017/11/21/rr-sress/. ŌĆó Mesirov, Jill P. ŌĆ£Accessible Reproducible Research.ŌĆØ Science, vol. 327, no. 5964, Jan. 2010, pp. 415ŌĆō16. science.sciencemag.org, doi:10.1126/science.1179653. ŌĆó Munaf├▓, Marcus R., et al. ŌĆ£A Manifesto for Reproducible Science.ŌĆØ Nature Human Behaviour, vol. 1, no. 1, Jan. 2017, p. 0021. www.nature.com, doi:10.1038/ s41562-016-0021. ŌĆó Patil, Prasad, et al. ŌĆ£A Statistical De’¼ünition for Reproducibility and Replicability.ŌĆØ BioRxiv, July 2016, p. 066803. www.biorxiv.org, doi:10.1101/066803. ŌĆó Russell, Pamela, et al. ŌĆ£A Large-Scale Analysis of Bioinformatics Code on GitHub.ŌĆØ BioRxiv, May 2018, p. 321919. www.biorxiv.org, doi:10.1101/321919. ŌĆó Schultheiss, Sebastian J., et al. ŌĆ£Persistence and Availability of Web Services in Computational Biology.ŌĆØ PLOS ONE, vol. 6, no. 9, Sept. 2011, p. e24914. PLoS Journals, doi:10.1371/journal.pone.0024914. 34
- 35. Stephen D. Turner, Ph.D. Bioinformatics Core Director University of Virginia School of Medicine bioinformatics.virginia.edu @strnr THANKYOU bit.ly/madssci2018repro doi.org/10.5281/zenodo.1255003


![We Are in the Middle of a
New Movement in Genomics
ŌĆó Genomics/bioinformatics advancing at grueling pace
- New questions
- New study designs
- New technologies, new [’¼üll-in-the-blank]-seq
ŌĆó New movements have:
- Leaders / method developers / early adopters
- First followers
- Everybody else
ŌĆó New technology leads to more reproducibility issues
2
CORES!](https://image.slidesharecdn.com/2018-06-20-abrf-rigor-reproducibility-190508205625/85/2018-ABRF-Tools-for-improving-rigor-and-reproducibility-in-bioinformatics-2-320.jpg)