AB-RNA-alignments-2010
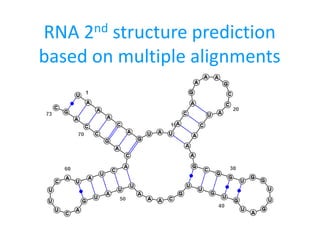
- 1. RNA 2nd structure prediction based on multiple alignments
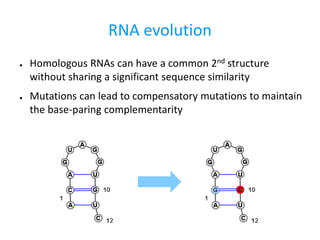
- 2. RNA evolution â Homologous RNAs can have a common 2nd structure without sharing a significant sequence similarity â Mutations can lead to compensatory mutations to maintain the base-paring complementarity
- 3. Comparative sequence analysis â In a structurally correct multiple alignment of RNAs, conserved base pairs are often revealed by the presence of frequent correlated compensatory mutations â Measure sequence covariation: mutual information â is the frequency of one of the four bases observed in col I â is the joint frequency of the base pairs observed in columns i and j ððð = â ðĨ ð,ðĨ ð ððĨ ð,ðĨ ð log2 ððĨ ð,ðĨ ð ððĨ ð â ððĨ ð ððĨ ð ððĨ ð,ðĨ ð
- 4. Covariance method G U C U U C G G A C G A C U U C G G U C G G C U U C G G C C ð2,9 = 3 â 1 3 â log2 1/3 1/9 = log23 â 1.59 ððð = â ðĨ ð,ðĨ ð ððĨ ð,ðĨ ð log2 ððĨ ð,ðĨ ð ððĨ ð â ððĨ ð â Mij varies between 0 and 2 â Mij is 2 when i and j appear completely random but are perfectly correlated â if i and j are uncorrelated, the mutual information is 0 â if either i or j are highly conserved positions, we also get little or no mutual information
- 5. â Mij is 2 when i and j appear completely random but are perfectly correlated â if i and j are uncorrelated, the mutual information is 0 â if either i or j are highly conserved positions, we also get little or no mutual information Covariance method G U C U U C G G A C G A C U U C G G U C G G C U U C G G C C G C C U U C G G G C ð1,9 = 4 â 1 4 â log2 1/4 1/4 = 0 ððð = â ðĨ ð,ðĨ ð ððĨ ð,ðĨ ð log2 ððĨ ð,ðĨ ð ððĨ ð â ððĨ ð ð2,9 = 4 â 1 4 â log2 1/4 1/16 = 2
- 6. Comparative analysis â Start with a multiple alignment â Predict 2nd structure base on alignment â Refine alignment based on 2nd structure â Repeat â The sequences to be compared must be sufficiently: â similar that they can be initially aligned by primary sequence â dissimilar that a number of covarying substitutions can be detected
- 7. Comparative analysis â How to build 2nd structure based on alignment? â Greedy method â choose the pair of columns that have the highest Mij â make a base pairs â carry on with the second highest Mij â problem columns might end up in more than one base pair
- 8. SCFGs and RNA alignments â An SCFG could be modified to generate columns of alignments instead of nucleotides â Requires a fixed number of sequences in the alignment â Instead, change it to generate the structure! ð â . ð âĢ ð. ð ðð Îĩ ð â ðð âĢ ðð âĢ ðð âĢ ðĒð ðð âĢ ðð âĢ ðð âĢ ððĒ ðððĒ âĢ ððð âĢ ðððĒ ðĒðð âĢ ððð âĢ ðĒðð ðð Îĩ
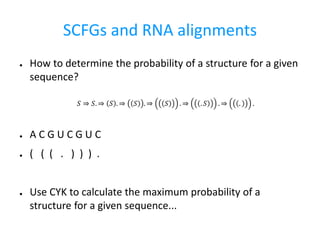
- 9. SCFGs and RNA alignments â How to determine the probability of a structure for a given sequence? â A C G U C G U C â ( ( ( . ) ) ) . â Use CYK to calculate the maximum probability of a structure for a given sequence... ð â ð. â ð . â ð . â ð . â . ð . â . .
- 10. SCFGs and RNA alignments â Use a phylogenetic tree (including branch lengths) to: â determine the probability of a column to be single â determine the probability of two columns to form a base pair â Use the SCFG and the columns probability to determine the best secondary structure for the alignment â CYK and the other SCFGs algorithms are basically the same
- 11. SCFGs and RNA alignments Knudsen&Hein 1999