AB-RNA-alignments-2011
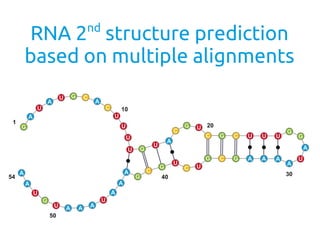
- 1. RNA 2nd structure prediction based on multiple alignments
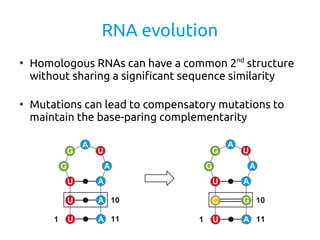
- 2. RNA evolution â Homologous RNAs can have a common 2nd structure without sharing a significant sequence similarity â Mutations can lead to compensatory mutations to maintain the base-paring complementarity
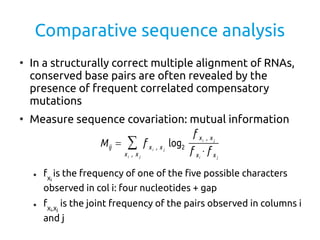
- 3. Comparative sequence analysis â In a structurally correct multiple alignment of RNAs, conserved base pairs are often revealed by the presence of frequent correlated compensatory mutations â Measure sequence covariation: mutual information â fXi is the frequency of one of the five possible characters observed in col i: four nucleotides + gap â fXi,Xj is the joint frequency of the pairs observed in columns i and j Mij = â xi , x j f xi , x j log2 f xi , x j f xi â f x j
- 4. Mutual information G U C U G G A C G A C U G G U C G G C U G G C C Mij = â xi , x j f xi , x j log2 f xi , x j f xi â f x j M2,7 = 3â (1 3 â log2 1/ 3 1/ 9)= log2 3 â 1.59 â Mij is maximum if i and j appear completely random but are perfectly correlated â if i and j are uncorrelated, the mutual information is 0 â if either i or j are highly conserved positions, we also get little or no mutual information
- 5. Mutual information â Mij is maximum if i and j appear completely random but are perfectly correlated â if i and j are uncorrelated, the mutual information is 0 â if either i or j are highly conserved positions, we also get little or no mutual information Mij = â xi , x j f xi , x j log2 f xi , x j f xi â f x j M2,7 = 4â (1 4 â log2 1 /4 1/16)= 2 M1,8 = log2 1 1 = 0 G U C U G G A C G A C U G G U C G G C U G G C C G C C U G G G C
- 6. Comparative analysis â Start with a multiple alignment â Predict 2nd structure base on alignment â Refine alignment based on 2nd structure â Repeat â The sequences to be compared must be sufficiently: â similar that they can be initially aligned by primary sequence â dissimilar that a number of co-varying substitutions can be detected
- 7. Comparative analysis â How to build 2nd structure based on alignment? â Greedy method â choose the pair of columns that have the highest Mij â make a base pair â carry on with the second highest Mij â Problem columns might end up in more than one base pair
- 8. Nussinov and alignments â Notations â aln the RNA alignment â alnk the kth sequence in the alignment â aln[i, j] the RNA alignment from position i to j â str the best 2nd structure for aln (over alphabet {(, ), .}) â str[i, j] the best2nd structure for aln[i, j] â score[i, j] the number of base pairs in str[i, j] â aln[i] · aln[j] if for all k, alnk [i] · alnk [j]
- 9. Nussinov and alignments â i unpaired and str[i+1, j] â j unpaired and str[i, j-1] â aln[i] · aln[j] and str[i+1, j-1] â str[i, k] and str[k+1, j] for some i < k < j i ji+1 i jj-1 i ji+1 j-1 i jk k+1
- 10. Nussinov and alignments â Scoring base pairs â on one sequence + 1 â on an alignment + 1 + Mij â Base pairs between columns with high mutual information are favoured â Other scoring schemes?
- 11. True Nussinov on alignment Nussinov single
- 12. From alignment structure to sequence structure A C G - - A A - U . . . . .(1 (2 )2 )1 A C G A A U . . . .(1 )1





![Nussinov and alignments
â
Notations
â
aln the RNA alignment
â alnk
the kth
sequence in the alignment
â
aln[i, j] the RNA alignment from position i to j
â
str the best 2nd
structure for aln
(over alphabet {(, ), .})
â
str[i, j] the best2nd
structure for aln[i, j]
â
score[i, j] the number of base pairs in str[i, j]
â aln[i] · aln[j] if for all k, alnk
[i] · alnk
[j]](https://image.slidesharecdn.com/36e98a6b-f4de-48a4-a25a-654d7ac26bf9-150826130457-lva1-app6891/85/AB-RNA-alignments-2011-8-320.jpg)
![Nussinov and alignments
â
i unpaired and str[i+1, j]
â
j unpaired and str[i, j-1]
â
aln[i] · aln[j] and str[i+1, j-1]
â
str[i, k] and str[k+1, j]
for some i < k < j
i ji+1
i jj-1
i ji+1 j-1
i jk k+1](https://image.slidesharecdn.com/36e98a6b-f4de-48a4-a25a-654d7ac26bf9-150826130457-lva1-app6891/85/AB-RNA-alignments-2011-9-320.jpg)


