AB-RNA-SCFGdesign=2010
- 2. SCFG design ● Dowell & Eddy (2004) G1: S  dS d ∣ d S ∣ S d ∣ SS ∣  G2: S  d S d ∣ d L ∣ Rd ∣ LS L  d S d ∣ aL R  Rd ∣  G3: S  d S ∣ d S d S ∣  G4: S  d S ∣ T ∣  T  T d ∣ d S d ∣ T d S d G5: S  LS ∣ L L  d F d ∣ d F  d F d ∣ LS
- 3. SCFG design ● Dowell & Eddy (2004) G1: S  dS d ∣ d S ∣ S d ∣ SS ∣  G2: S  d S d ∣ d L ∣ Rd ∣ LS L  d S d ∣ aL R  Rd ∣  G3: S  d S ∣ d S d S ∣  G4: S  d S ∣ T ∣  T  T d ∣ d S d ∣ T d S d G5: S  LS ∣ L L  d F d ∣ d F  d F d ∣ LS
- 4. Search for better SCFGs ‚óè Evolutionary algorithm ‚óè Initial population ‚óè Mutation model ‚óè Breeding model ‚óè Selection
- 5. Normal form ● Chomsky Normal Form ● CNF makes it hard to determine the associated structure ● Use a different normal form A  BC A  d A  d B d A  BC A  d A  
- 6. CYK V x Vy Vz i k k+1 j score[x ,i , j]= { 0 if ji ex , seq[i] if i=j max i≤kj V x V y V z score[y ,i ,k]⋅score[z ,k1, j]⋅tx , y , z
- 7. CYK V x Vy Vz i k k+1 j V x Vy i+1 j -1 i j score[x ,i , j]= { 0 if ji ex , seq[i] if i=j max i≤kj V x V y V z score[y ,i ,k]⋅score[z ,k1, j]⋅tx , y , z
- 8. CYK V x Vy Vz i k k+1 j V x Vy i+1 j -1 i j score[x ,i , j]= { 0 if ji ex , seq[i] if i= j max { max i≤k j V x Vy V z score[y ,i ,k]⋅score[z ,k1, j]⋅tx , y , z max V x d V y d score[y ,i1, j−1]⋅wx , y ,d , d⋅1seq[i]=d∧seq[ j]=d
- 9. CYK score[x ,i , j]= { 0 if ji ex , seq[i] if i= j max { max i≤k j V x Vy V z score[y ,i ,k]⋅score[z ,k1, j]⋅tx , y , z max V x d V y d score[y ,i1, j−1]⋅wx , y ,d , d⋅1seq[i]=d∧seq[ j]=d score[x ,i , j]= { 0 if ji ex , seq[i] if i=j max i≤kj V x V y V z score[y ,i ,k]⋅score[z ,k1, j]⋅tx , y , z
- 10. Initial population ‚óè Use grammars from Dowell&Eddy ‚óè biased search ‚óè Use random grammars ‚óè hard to generate random grammars ‚óè Use small grammars with only 2 nonterminals
- 11. Mutations ‚óè Insert rule ‚óè Delete rule ‚óè Modify rule ‚óè Modify start variable ‚óè Add nonterminal + 2 new rules ‚óè Simulate rule A ‚Üí B
- 12. Mutations ‚óè Insert rule ‚óè Delete rule ‚óè Modify rule ‚óè Modify start variable ‚óè Add nonterminal + 2 new rules ‚óè Simulate rule A ‚Üí B ‚óè Problem with what probabilities should these mutations occur?
- 13. Mutations ‚óè Insert rule 4 / 13 ‚óè Delete rule 1 / 13 ‚óè Modify rule 2 / 13 ‚óè Modify start variable 1 / 13 ‚óè Add nonterminal + 2 new rules 4 / 13 ‚óè Simulate rule A ‚Üí B 1 / 13 Everything else: uniform ‚óè Problem with what probabilities should these mutations occur?
- 14. Breeding ‚óè Captivate characteristics of two grammars in one Initial grammars: ‚óè G1 with nonterminals V1 , V2 , ..., Vn ‚óè G2 with nonterminals W1 , W2 , ..., Wm Bred grammar: ‚óè G with nonterminals S, V2 , ..., Vn , W2 , ..., Wm ‚óè The set of rules for G is simply the union from G1 and G2 with V1 and W1 replaced by S ‚óè Properties
- 15. Decisions, decisions... ● How to select the grammars to mutate? ● How to select the grammars to breed? ● depending on the fitness of the grammar – just one mutation or breeding is unlikely to improve fitness – could bias the search ● independent of the fitness – escape local optimum
- 16. Selection ● Fitness: ● RNA 2nd structure metrics ● sensitivity and specificity ● Using fitness ● select deterministically ● select probabilistically ● Next generation ● mutated/bred grammars – better for escaping local optimum ● both “old” grammars and mutated/bred grammars
- 17. RNA metrics S1 ( . . . . . . ) S2 . . ( . . ) . . S . . . . . . . . distance(S1, S) = distance (S2, S) ? distance(S1, S) ≠ distance (S2, S) ?
- 18. RNA metrics S1 ( . . . . . . ) S2 . . ( . . ) . . S . . . . . . . . distance(S1, S) = distance (S2, S) ? distance(S1, S) ≠ distance (S2, S) ? ● Different metrics – different characteristics
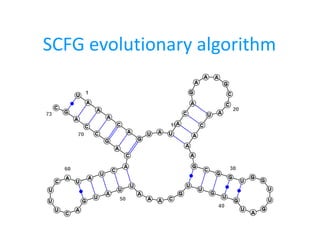
- 19. Results G0: A  BA ∣ d ∣ d C d B  d ∣ d C d C  BA ∣ d C d G5: A  CC ∣ CB ∣ BC ∣ EC ∣ d A d ∣ d E d B  d C  CD ∣ Bb ∣ d A d D  GC ∣ d C d E  AB ∣ CD ∣ d F  AB G  FB G7: A  DE ∣ AB ∣ BA ∣ AH ∣ d ∣ d F d ∣ d H d B  d C  d H d D  BB ∣ AC E  d ∣ d H d F  FB ∣ CF ∣ d G  GH ∣ d H d ∣ dC d H  FA ∣ AF ∣ HH ∣ d B ∣ d H d Gram Dowell&Eddy RNA strand sens spec sens spec G0 0.465 0.406 0.496 0.479 G5 0.487 0.432 0.469 0.467 G7 0.465 0.376 0.526 0.479
- 20. Results G0: A  BA ∣ d ∣ d C d B  d ∣ d C d C  BA ∣ d C d G5: A  CC ∣ CB ∣ BC ∣ EC ∣ d A d ∣ d E d B  d C  CD ∣ Bb ∣ d A d D  GC ∣ d C d E  AB ∣ CD ∣ d F  AB G  FB G7: A  DE ∣ AB ∣ BA ∣ AH ∣ d ∣ d F d ∣ d H d B  d C  d H d D  BB ∣ AC E  d ∣ d H d F  FB ∣ CF ∣ d G  GH ∣ d H d ∣ dC d H  FA ∣ AF ∣ HH ∣ d B ∣ d H d Gram Dowell&Eddy RNA strand sens spec sens spec G0 0.465 0.406 0.496 0.479 G5 0.487 0.432 0.469 0.467 G7 0.465 0.376 0.526 0.479 D&E data contains pseudoknots!





![CYK
V x
Vy Vz
i k k+1 j
score[x ,i , j]=
{
0 if jÓÇÑi
eÓÇûx , seq[i]ÓÇü if i=j
max
i≤kj
V x ÓÇåV y V z
score[y ,i ,k]‚ãÖscore[z ,kÓÇÉ1, j]‚ãÖtÓÇûx , y , zÓÇü](https://image.slidesharecdn.com/2cfcf06a-b202-44c4-8d0b-8e49e251bc9b-150826130144-lva1-app6891/85/AB-RNA-SCFGdesign-2010-6-320.jpg)
![CYK
V x
Vy Vz
i k k+1 j
V x
Vy
i+1 j -1
i j
score[x ,i , j]=
{
0 if jÓÇÑi
eÓÇûx , seq[i]ÓÇü if i=j
max
i≤kj
V x ÓÇåV y V z
score[y ,i ,k]‚ãÖscore[z ,kÓÇÉ1, j]‚ãÖtÓÇûx , y , zÓÇü](https://image.slidesharecdn.com/2cfcf06a-b202-44c4-8d0b-8e49e251bc9b-150826130144-lva1-app6891/85/AB-RNA-SCFGdesign-2010-7-320.jpg)
![CYK
V x
Vy Vz
i k k+1 j
V x
Vy
i+1 j -1
i j
score[x ,i , j]=
{
0 if jÓÇÑi
eÓÇûx , seq[i]ÓÇü if i= j
max
{
max
i≤k j
V x ÓÇåVy V z
score[y ,i ,k]‚ãÖscore[z ,kÓÇÉ1, j]‚ãÖtÓÇûx , y , zÓÇü
max
V x ÓÇåd V y
ÓÇëd
score[y ,iÓÇÉ1, j‚àí1]‚ãÖwÓÇûx , y ,d , ÓÇëdÓÇü‚ãÖ1seq[i]=d‚àßseq[ j]=ÓÇëd](https://image.slidesharecdn.com/2cfcf06a-b202-44c4-8d0b-8e49e251bc9b-150826130144-lva1-app6891/85/AB-RNA-SCFGdesign-2010-8-320.jpg)
![CYK
score[x ,i , j]=
{
0 if jÓÇÑi
eÓÇûx , seq[i]ÓÇü if i= j
max
{
max
i≤k j
V x ÓÇåVy V z
score[y ,i ,k]‚ãÖscore[z ,kÓÇÉ1, j]‚ãÖtÓÇûx , y , zÓÇü
max
V x ÓÇåd V y
ÓÇëd
score[y ,iÓÇÉ1, j‚àí1]‚ãÖwÓÇûx , y ,d , ÓÇëdÓÇü‚ãÖ1seq[i]=d‚àßseq[ j]=ÓÇëd
score[x ,i , j]=
{
0 if jÓÇÑi
eÓÇûx , seq[i]ÓÇü if i=j
max
i≤kj
V x ÓÇåV y V z
score[y ,i ,k]‚ãÖscore[z ,kÓÇÉ1, j]‚ãÖtÓÇûx , y , zÓÇü](https://image.slidesharecdn.com/2cfcf06a-b202-44c4-8d0b-8e49e251bc9b-150826130144-lva1-app6891/85/AB-RNA-SCFGdesign-2010-9-320.jpg)










