Comparison of Genomic DNA to cDNA Alignment Methods
Download as ppt, pdf3 likes770 views
This document compares different genomic DNA alignment methods, including a classic semi-global algorithm and heuristic tools like sim4 and Spidey. It analyzes the performance of these aligners on subsets of human gene and mRNA data, evaluating metrics like number of gaps, exon differences, sequence similarity, and runtime. The results show that the classic semi-global aligner produces very high-quality alignments but is slow, while sim4 achieves good accuracy faster than other external tools and outperforms them on most metrics.
1 of 19
Downloaded 11 times









Ad
Recommended
qPCR Design Strategies for Specific Applications
qPCR Design Strategies for Specific ApplicationsIntegrated DNA Technologies
╠²
This document provides guidance on designing quantitative PCR (qPCR) assays for specific applications, including species-specific, strain-specific, and copy number variation (CNV) assays. It outlines general design strategies and considerations, including using sequence alignments to identify unique target regions and primers that avoid nonspecific amplification. Examples are provided for designing assays to distinguish similar genes in Arabidopsis thaliana and related viral strains. Design of CNV assays is also discussed, highlighting the importance of a single-copy reference gene.Dna library lecture-Gene libraries and screening
Dna library lecture-Gene libraries and screening Abdullah Abobakr
╠²
This document discusses gene libraries and screening procedures. It begins by explaining what genomic and cDNA libraries are. It then provides details on creating genomic libraries, including purifying genomic DNA, fragmenting it, and cloning the fragments into vectors. Creating cDNA libraries involves isolating mRNA, synthesizing cDNA, and ligating the cDNA to vectors. The size of libraries needed to ensure coverage of genomes is calculated. Lambda phage is described as a commonly used vector that can accept inserts up to 23kb in size. The processes of packaging recombinant DNA into lambda phage particles and creating lambda phage libraries are outlined.AlgoAlignementGenomicSequences.ppt
AlgoAlignementGenomicSequences.pptSkanderBena
╠²
This document summarizes an academic presentation about algorithms for aligning genomic sequences. It discusses edit distance models for comparing sequences and finding the best alignment. Local, global, and glocal alignment algorithms are introduced, including CHAOS for local alignment and LAGAN and Shuffle-LAGAN for multiple global and glocal alignments of whole genomes. Applications to aligning the cystic fibrosis gene and analyzing rearrangements between human and mouse are also summarized.GPCODON ALIGNMENT: A GLOBAL PAIRWISE CODON BASED SEQUENCE ALIGNMENT APPROACH
GPCODON ALIGNMENT: A GLOBAL PAIRWISE CODON BASED SEQUENCE ALIGNMENT APPROACHIJDMS
╠²
This document introduces a global pairwise DNA sequence alignment method called gpcodon alignment, which employs a new empirical codon substitution scoring matrix for improved accuracy. The paper discusses the limitations of traditional sequence alignment techniques and presents experimental results demonstrating the efficiency and effectiveness of the proposed approach over a comparable tool. Various algorithms and previous works relevant to sequence alignment are also reviewed, laying the groundwork for the proposed methodology.Virus Sequence Alignment and Phylogenetic Analysis 2019
Virus Sequence Alignment and Phylogenetic Analysis 2019Bioinformatics and Computational Biosciences Branch
╠²
This document discusses phylogenetic analysis and tree building. It introduces the Bioinformatics and Computational Biology Branch (BCBB) group and their work analyzing biological sequences and constructing phylogenetic trees. The document explains why biological sequences are important to analyze and compares sequences to understand relatedness and evolution. It also covers multiple sequence alignment, substitution models, and algorithms for building trees, including neighbor-joining.Aug2013 NIST highly confident genotype calls for NA12878
Aug2013 NIST highly confident genotype calls for NA12878GenomeInABottle
╠²
This document discusses the creation of a set of highly confident genotypes for the genome NA12878 by integrating data from 12 datasets across 5 sequencing platforms. The goals were to define regions of the genome with no false positive or negative genotype calls, include as much of the genome as possible, and avoid biases. Candidate variants were identified and datasets were removed that showed characteristics of sequencing, mapping, or alignment biases until all datasets agreed. Regions like repeats and duplications were labeled uncertain. Verification showed the calls were highly accurate except for some systematic errors and complex variants. The data and calls are available to help assess performance of variant calling.Sequence alignment
Sequence alignmentDr. Harisingh Gour Vishwavidyalaya (A Central Universuty), Sagar, MP
╠²
This document discusses sequence alignment, which involves arranging biological sequences like DNA, RNA, or proteins to identify regions of similarity. It covers the basic concepts of sequence alignment including global versus local alignment and different methods like dot matrix, dynamic programming, and word-based approaches. Dynamic programming is highlighted as the most common algorithm that uses a scoring system to find the optimal alignment between two sequences.BLAST and sequence alignment
BLAST and sequence alignmentBioinformatics and Computational Biosciences Branch
╠²
This document provides an overview of pairwise sequence alignment and BLAST. It discusses how pairwise alignment works using substitution matrices to assign homology between sites. It demonstrates the dynamic programming approach to pairwise alignment calculation and describes how local alignments are identified. The document also introduces BLAST and how it uses word matching to rapidly identify similar sequences in a database and then performs local alignments on matching regions.Phylogenetics1
Phylogenetics1S├®bastien De Landtsheer
╠²
The document summarizes an introduction to phylogenetics workshop covering DNA alignments, distance matrices, and distance-based tree inference methods. Part I of the workshop introduces phylogenetics and classification of species, describes DNA alignment algorithms like Needleman-Wunsch, and explains distance-based tree building methods like UPGMA and Neighbor-Joining that construct phylogenetic trees from distance matrices. Software for alignments, distance calculations, and tree building is also listed.Implementation of DNA sequence alignment algorithms using Fpga ,ML,and CNN
Implementation of DNA sequence alignment algorithms using Fpga ,ML,and CNNAmr Rashed
╠²
The document discusses implementing DNA/RNA sequence alignment algorithms using FPGA. It proposes a hardware implementation that relies on complete parallelization of algorithms like Smith-Waterman and Needleman-Wunsch under certain limitations. A lookup table is used to accelerate the algorithms in O(N/4) time. Fifty-four Boolean functions are derived for parallel implementation. The implementation represents DNA/RNA sequences as combinations of 4 nucleotides and is applicable to both software and hardware.06_Alignment_2022.pdf
06_Alignment_2022.pdfKristen DeAngelis
╠²
This document discusses sequence alignment and its applications in bioinformatics. It begins by explaining the goals of learning about homology and how sequence alignment relates to function across organisms. It then describes different types of sequence alignment including global, local, Needleman-Wunsch, Smith-Waterman, and BLAST. It explains how to quantify alignment scores and perform statistical analysis of alignments. The document provides examples of alignment matrices and algorithms for finding the best alignment between sequences.Snippy - Rapid bacterial variant calling - UK - tue 5 may 2015
Snippy - Rapid bacterial variant calling - UK - tue 5 may 2015Torsten Seemann
╠²
The document outlines rapid bacterial variant calling and core genome alignments, detailing the capabilities of tools like Snippy for identifying genetic variations such as SNPs and indels from bacterial genomes. It also discusses the transition of microbiological services towards modern genomic techniques under Professor Ben Howden's leadership at the oldest public health lab in Australia. Additionally, the document summarizes the challenges and methodologies in phylogenetics and whole genome alignment, emphasizing the utility and speed of Snippy in processing genomic data.Under the Hood of Alignment Algorithms for NGS Researchers
Under the Hood of Alignment Algorithms for NGS Researchers
Golden Helix Inc
╠²
The document discusses alignment algorithms for next-generation sequencing (NGS) researchers, focusing on the importance of correctly aligning genomic reads to reference sequences. It outlines different methods for alignment, including dynamic programming and hash-based approaches, and compares the efficiency of various algorithms like BWA, Bowtie, and others. Additionally, it highlights challenges in variant calling and mapping quality, emphasizing the need for accurate algorithms to handle complex genomic data.ą┐čĆąĄąĘąĄąĮčéą░čåąĖčÅ ąĘą░ ą▓ą░čĆčłą░ą▓ą░
ą┐čĆąĄąĘąĄąĮčéą░čåąĖčÅ ąĘą░ ą▓ą░čĆčłą░ą▓ą░Valeriya Simeonova
╠²
An approach is developed to detect and correct errors in 16S RNA fragments from metagenomic sequencing data. Two algorithms are proposed - the first finds and corrects errors by studying correspondence between similar sequences, while the second fine-tunes the first algorithm's accuracy for estimating sequence errors, SNPs and detecting species. The approaches are tested on two 16S RNA fragment datasets, and classification results after error correction are compared to evaluate performance. Future work includes improving error detection and correction and validating the approach on other datasets.Bioalgo 2012-01-gene-prediction-sim
Bioalgo 2012-01-gene-prediction-simBioinformaticsInstitute
╠²
Similarity-based gene prediction approaches compare unknown genes in a genome to known genes in closely-related species to determine gene structure and function. A key challenge is that genes are split into exons and introns. Spliced alignment algorithms aim to find the best chain of exons in the genome that align to a target gene sequence, taking into account local alignments between potential exons. Popular gene prediction tools that use spliced alignment include GENSCAN, GenomeScan, TwinScan, Glimmer, and GenMark. They apply probabilistic models and similarity information to predict exon-intron structure.Molecular Subtyping of Breast Cancer and Somatic Mutation Discovery Using DNA...
Molecular Subtyping of Breast Cancer and Somatic Mutation Discovery Using DNA...Setia Pramana
╠²
The document discusses molecular subtyping of breast cancer and somatic mutation discovery using DNA and RNA sequencing, highlighting the complexities and heterogeneity of breast cancer subtypes. It emphasizes the role of somatic mutations in cancer development and the use of next-generation sequencing (NGS) to enhance understanding and treatment decisions for breast cancer. The study aims to identify new biomarkers and therapeutic targets through the analysis of genetic data from breast cancer samples.Comparative genomics
Comparative genomicsAthira RG
╠²
Comparative genomics involves systematically comparing genome sequences from different organisms. It uses computer programs to identify homologous genomic regions and align sequences at the base-pair level. Comparing genomes at different phylogenetic distances can provide insights into gene structure/function, evolution, and characteristics unique to each organism. Key tools for comparative genomics include genome browsers, aligners, and databases that classify orthologous gene clusters conserved across species.Msa & rooted/unrooted tree
Msa & rooted/unrooted treeSamiul Ehsan
╠²
Phylogenetics is the study of evolutionary history and relationships between taxa. Phylogenetic trees present relationships as a collection of nodes and branches, with closely related taxa appearing near each other. Multiple sequence alignment (MSA) is used to reveal biological facts about sequences and to construct phylogenetic trees. However, MSA is computationally complex due to the exponential growth in possible alignments as more sequences are added.20100515 bioinformatics kapushesky_lecture07
20100515 bioinformatics kapushesky_lecture07Computer Science Club
╠²
The document discusses pairwise sequence alignment methods. It defines key concepts like homology and orthology. It explains that dynamic programming is used to find optimal alignments through building a score matrix and backtracking. Global alignment finds the best match over full sequences while local alignment identifies regions of local similarity. Scoring systems like PAM matrices assign values based on substitutions and penalties for gaps.Comparitive genomics
Comparitive genomicsInternational Islamic University Islamabad
╠²
Comparative genomics allows scientists to compare the genomes of different organisms to better understand genome structure and function. By aligning homologous genes, researchers can identify regions of similarity and difference between species that provide insights into evolution, gene function, and disease. The Ensembl database facilitates comparative genomic analyses across 69 vertebrate and eukaryotic species by allowing users to search for similar sequences, view alignments and gene trees, and integrate external genomic data. Comparisons between human and mouse genomes reveal approximately 85% identity in coding regions and gene structures but more divergence in intron sequences and lengths.Blast fasta 4
Blast fasta 4Er Puspendra Tripathi
╠²
BLAST and FASTA are algorithms for searching sequence databases to find local alignments between a query sequence and database sequences, with BLAST providing faster searches and improved statistical analysis compared to FASTA. Both algorithms work by first identifying short exact matches between sequences and then extending these matches to identify longer regions of similarity. The algorithms model DNA and protein sequence alignments as coin tosses to determine the expected length of the longest matching region between random sequences.Gene targeting and sequence tags
Gene targeting and sequence tagsAlen Shaji
╠²
The document discusses gene targeting, a method to modify specific genes using a targeting vector and homologous recombination, enabling gene knockout or introduction for research and therapeutic purposes. It also details expressed sequence tags (ESTs), which are short sequences used in gene discovery and mapping, with implications for understanding gene expression and function. The significance of the work in gene targeting is highlighted by the 2007 Nobel Prize awarded to key researchers in this field.Processamento de tweets em tempo real com Python, Django e Celery - TDC 2014Miguel Galves
╠²
O documento descreve o lan├¦amento de uma ferramenta chamada Tuit├┤metro que permite acompanhar em tempo real a discuss├Żo sobre futebol brasileiro no Twitter. A ferramenta mede a "empolga├¦├Żo" dos torcedores com base na quantidade e assuntos dos tweets. O sistema faz o processamento dos tweets em tempo real usando Django, Celery e Redis.Redis para iniciantes - TDC 2014Miguel Galves
╠²
O documento apresenta Redis, um banco de dados chave-valor em mem├│ria, e discute como ele pode ser usado para cache, gerenciamento de sess├Żo, filas e outras aplica├¦├Ąes. Ele explica como instalar e usar Redis, seus tipos de dados e comandos, e d├Ī exemplos de como empresas grandes como Twitter e GitHub usam Redis em produ├¦├Żo.New Strategy to detect SNPs
New Strategy to detect SNPsMiguel Galves
╠²
This document describes a new strategy for detecting single nucleotide polymorphisms (SNPs) in HIV genetic sequences. The strategy involves trimming low quality bases, correcting base calls using area ratio or average height ratio algorithms, filtering adjacent SNPs, and generating a consensus sequence from multiple alignments. The strategy was tested on 35 batches of HIV sequences and achieved true positive rates of 75.4% using area ratio and 52.6% using average height ratio, outperforming two external SNP detection tools, Polybayes and Polyphred. Future work will involve testing with larger datasets with higher coverage and sequences from more conserved organisms.Qualifica├¦├Żo de MestradoMiguel Galves
╠²
Este documento descreve uma abordagem computacional para o estudo de polimorfismos de base ├║nica (SNPs). Ele discute as etapas do processo, incluindo alinhamento de sequ├¬ncias, detec├¦├Żo de SNPs e medi├¦├Żo da correla├¦├Żo entre SNPs. O objetivo ├® desenvolver novas metodologias para cada etapa e integr├Ī-las em um sistema de identifica├¦├Żo de SNPs.Uma abordagem computacional para a determina├¦├Żo de polimorfismos de base ├║nicaMiguel Galves
╠²
Este documento resume uma abordagem computacional para determina├¦├Żo de polimorfismos de base ├║nica (SNPs) atrav├®s de tr├¬s etapas: alinhamento de seq├╝├¬ncias, detec├¦├Żo de SNPs em cromatogramas e an├Īlise de correla├¦├Ąes entre SNPs. Os resultados obtidos nas tr├¬s etapas foram satisfat├│rios e demonstraram a viabilidade da abordagem proposta.Django: Uso de frameworks ├Īgeis para desenvolvimento webMiguel Galves
╠²
O documento discute o framework Django para desenvolvimento web, comparando-o com J2EE. Django ├® mais simples e focado em produtividade, enquanto J2EE ├® mais complexo. Embora linguagens din├ómicas como Python possam ter desempenho inferior, frameworks como Django permitem desenvolver aplica├¦├Ąes escal├Īveis com o suporte de ferramentas externas como caches e balanceadores de carga.GIS em 3 horasMiguel Galves
╠²
O documento fornece uma introdu├¦├Żo sobre Sistemas de Informa├¦├Żo Geogr├Īfica (SIG), explicando conceitos b├Īsicos como georreferenciamento, modelos vetoriais e raster, proje├¦├Ąes cartogr├Īficas, armazenamento de dados espaciais e protocolos de acesso como WMS e WFS.AJAXMiguel Galves
╠²
O documento apresenta um hist├│rico da web, como evoluiu a arquitetura de sites e a introdu├¦├Żo do AJAX. Explica que AJAX utiliza tecnologias j├Ī existentes como JavaScript, XML e HTTP para permitir atualiza├¦├Ąes ass├Łncronas na p├Īgina e maior interatividade. Tamb├®m destaca os desafios do desenvolvimento com AJAX, como compatibilidade entre navegadores e a necessidade de implementar mecanismos como hist├│rico de navega├¦├Żo.More Related Content
Similar to Comparison of Genomic DNA to cDNA Alignment Methods (14)
Phylogenetics1
Phylogenetics1S├®bastien De Landtsheer
╠²
The document summarizes an introduction to phylogenetics workshop covering DNA alignments, distance matrices, and distance-based tree inference methods. Part I of the workshop introduces phylogenetics and classification of species, describes DNA alignment algorithms like Needleman-Wunsch, and explains distance-based tree building methods like UPGMA and Neighbor-Joining that construct phylogenetic trees from distance matrices. Software for alignments, distance calculations, and tree building is also listed.Implementation of DNA sequence alignment algorithms using Fpga ,ML,and CNN
Implementation of DNA sequence alignment algorithms using Fpga ,ML,and CNNAmr Rashed
╠²
The document discusses implementing DNA/RNA sequence alignment algorithms using FPGA. It proposes a hardware implementation that relies on complete parallelization of algorithms like Smith-Waterman and Needleman-Wunsch under certain limitations. A lookup table is used to accelerate the algorithms in O(N/4) time. Fifty-four Boolean functions are derived for parallel implementation. The implementation represents DNA/RNA sequences as combinations of 4 nucleotides and is applicable to both software and hardware.06_Alignment_2022.pdf
06_Alignment_2022.pdfKristen DeAngelis
╠²
This document discusses sequence alignment and its applications in bioinformatics. It begins by explaining the goals of learning about homology and how sequence alignment relates to function across organisms. It then describes different types of sequence alignment including global, local, Needleman-Wunsch, Smith-Waterman, and BLAST. It explains how to quantify alignment scores and perform statistical analysis of alignments. The document provides examples of alignment matrices and algorithms for finding the best alignment between sequences.Snippy - Rapid bacterial variant calling - UK - tue 5 may 2015
Snippy - Rapid bacterial variant calling - UK - tue 5 may 2015Torsten Seemann
╠²
The document outlines rapid bacterial variant calling and core genome alignments, detailing the capabilities of tools like Snippy for identifying genetic variations such as SNPs and indels from bacterial genomes. It also discusses the transition of microbiological services towards modern genomic techniques under Professor Ben Howden's leadership at the oldest public health lab in Australia. Additionally, the document summarizes the challenges and methodologies in phylogenetics and whole genome alignment, emphasizing the utility and speed of Snippy in processing genomic data.Under the Hood of Alignment Algorithms for NGS Researchers
Under the Hood of Alignment Algorithms for NGS Researchers
Golden Helix Inc
╠²
The document discusses alignment algorithms for next-generation sequencing (NGS) researchers, focusing on the importance of correctly aligning genomic reads to reference sequences. It outlines different methods for alignment, including dynamic programming and hash-based approaches, and compares the efficiency of various algorithms like BWA, Bowtie, and others. Additionally, it highlights challenges in variant calling and mapping quality, emphasizing the need for accurate algorithms to handle complex genomic data.ą┐čĆąĄąĘąĄąĮčéą░čåąĖčÅ ąĘą░ ą▓ą░čĆčłą░ą▓ą░
ą┐čĆąĄąĘąĄąĮčéą░čåąĖčÅ ąĘą░ ą▓ą░čĆčłą░ą▓ą░Valeriya Simeonova
╠²
An approach is developed to detect and correct errors in 16S RNA fragments from metagenomic sequencing data. Two algorithms are proposed - the first finds and corrects errors by studying correspondence between similar sequences, while the second fine-tunes the first algorithm's accuracy for estimating sequence errors, SNPs and detecting species. The approaches are tested on two 16S RNA fragment datasets, and classification results after error correction are compared to evaluate performance. Future work includes improving error detection and correction and validating the approach on other datasets.Bioalgo 2012-01-gene-prediction-sim
Bioalgo 2012-01-gene-prediction-simBioinformaticsInstitute
╠²
Similarity-based gene prediction approaches compare unknown genes in a genome to known genes in closely-related species to determine gene structure and function. A key challenge is that genes are split into exons and introns. Spliced alignment algorithms aim to find the best chain of exons in the genome that align to a target gene sequence, taking into account local alignments between potential exons. Popular gene prediction tools that use spliced alignment include GENSCAN, GenomeScan, TwinScan, Glimmer, and GenMark. They apply probabilistic models and similarity information to predict exon-intron structure.Molecular Subtyping of Breast Cancer and Somatic Mutation Discovery Using DNA...
Molecular Subtyping of Breast Cancer and Somatic Mutation Discovery Using DNA...Setia Pramana
╠²
The document discusses molecular subtyping of breast cancer and somatic mutation discovery using DNA and RNA sequencing, highlighting the complexities and heterogeneity of breast cancer subtypes. It emphasizes the role of somatic mutations in cancer development and the use of next-generation sequencing (NGS) to enhance understanding and treatment decisions for breast cancer. The study aims to identify new biomarkers and therapeutic targets through the analysis of genetic data from breast cancer samples.Comparative genomics
Comparative genomicsAthira RG
╠²
Comparative genomics involves systematically comparing genome sequences from different organisms. It uses computer programs to identify homologous genomic regions and align sequences at the base-pair level. Comparing genomes at different phylogenetic distances can provide insights into gene structure/function, evolution, and characteristics unique to each organism. Key tools for comparative genomics include genome browsers, aligners, and databases that classify orthologous gene clusters conserved across species.Msa & rooted/unrooted tree
Msa & rooted/unrooted treeSamiul Ehsan
╠²
Phylogenetics is the study of evolutionary history and relationships between taxa. Phylogenetic trees present relationships as a collection of nodes and branches, with closely related taxa appearing near each other. Multiple sequence alignment (MSA) is used to reveal biological facts about sequences and to construct phylogenetic trees. However, MSA is computationally complex due to the exponential growth in possible alignments as more sequences are added.20100515 bioinformatics kapushesky_lecture07
20100515 bioinformatics kapushesky_lecture07Computer Science Club
╠²
The document discusses pairwise sequence alignment methods. It defines key concepts like homology and orthology. It explains that dynamic programming is used to find optimal alignments through building a score matrix and backtracking. Global alignment finds the best match over full sequences while local alignment identifies regions of local similarity. Scoring systems like PAM matrices assign values based on substitutions and penalties for gaps.Comparitive genomics
Comparitive genomicsInternational Islamic University Islamabad
╠²
Comparative genomics allows scientists to compare the genomes of different organisms to better understand genome structure and function. By aligning homologous genes, researchers can identify regions of similarity and difference between species that provide insights into evolution, gene function, and disease. The Ensembl database facilitates comparative genomic analyses across 69 vertebrate and eukaryotic species by allowing users to search for similar sequences, view alignments and gene trees, and integrate external genomic data. Comparisons between human and mouse genomes reveal approximately 85% identity in coding regions and gene structures but more divergence in intron sequences and lengths.Blast fasta 4
Blast fasta 4Er Puspendra Tripathi
╠²
BLAST and FASTA are algorithms for searching sequence databases to find local alignments between a query sequence and database sequences, with BLAST providing faster searches and improved statistical analysis compared to FASTA. Both algorithms work by first identifying short exact matches between sequences and then extending these matches to identify longer regions of similarity. The algorithms model DNA and protein sequence alignments as coin tosses to determine the expected length of the longest matching region between random sequences.Gene targeting and sequence tags
Gene targeting and sequence tagsAlen Shaji
╠²
The document discusses gene targeting, a method to modify specific genes using a targeting vector and homologous recombination, enabling gene knockout or introduction for research and therapeutic purposes. It also details expressed sequence tags (ESTs), which are short sequences used in gene discovery and mapping, with implications for understanding gene expression and function. The significance of the work in gene targeting is highlighted by the 2007 Nobel Prize awarded to key researchers in this field.More from Miguel Galves (9)
Processamento de tweets em tempo real com Python, Django e Celery - TDC 2014Miguel Galves
╠²
O documento descreve o lan├¦amento de uma ferramenta chamada Tuit├┤metro que permite acompanhar em tempo real a discuss├Żo sobre futebol brasileiro no Twitter. A ferramenta mede a "empolga├¦├Żo" dos torcedores com base na quantidade e assuntos dos tweets. O sistema faz o processamento dos tweets em tempo real usando Django, Celery e Redis.Redis para iniciantes - TDC 2014Miguel Galves
╠²
O documento apresenta Redis, um banco de dados chave-valor em mem├│ria, e discute como ele pode ser usado para cache, gerenciamento de sess├Żo, filas e outras aplica├¦├Ąes. Ele explica como instalar e usar Redis, seus tipos de dados e comandos, e d├Ī exemplos de como empresas grandes como Twitter e GitHub usam Redis em produ├¦├Żo.New Strategy to detect SNPs
New Strategy to detect SNPsMiguel Galves
╠²
This document describes a new strategy for detecting single nucleotide polymorphisms (SNPs) in HIV genetic sequences. The strategy involves trimming low quality bases, correcting base calls using area ratio or average height ratio algorithms, filtering adjacent SNPs, and generating a consensus sequence from multiple alignments. The strategy was tested on 35 batches of HIV sequences and achieved true positive rates of 75.4% using area ratio and 52.6% using average height ratio, outperforming two external SNP detection tools, Polybayes and Polyphred. Future work will involve testing with larger datasets with higher coverage and sequences from more conserved organisms.Qualifica├¦├Żo de MestradoMiguel Galves
╠²
Este documento descreve uma abordagem computacional para o estudo de polimorfismos de base ├║nica (SNPs). Ele discute as etapas do processo, incluindo alinhamento de sequ├¬ncias, detec├¦├Żo de SNPs e medi├¦├Żo da correla├¦├Żo entre SNPs. O objetivo ├® desenvolver novas metodologias para cada etapa e integr├Ī-las em um sistema de identifica├¦├Żo de SNPs.Uma abordagem computacional para a determina├¦├Żo de polimorfismos de base ├║nicaMiguel Galves
╠²
Este documento resume uma abordagem computacional para determina├¦├Żo de polimorfismos de base ├║nica (SNPs) atrav├®s de tr├¬s etapas: alinhamento de seq├╝├¬ncias, detec├¦├Żo de SNPs em cromatogramas e an├Īlise de correla├¦├Ąes entre SNPs. Os resultados obtidos nas tr├¬s etapas foram satisfat├│rios e demonstraram a viabilidade da abordagem proposta.Django: Uso de frameworks ├Īgeis para desenvolvimento webMiguel Galves
╠²
O documento discute o framework Django para desenvolvimento web, comparando-o com J2EE. Django ├® mais simples e focado em produtividade, enquanto J2EE ├® mais complexo. Embora linguagens din├ómicas como Python possam ter desempenho inferior, frameworks como Django permitem desenvolver aplica├¦├Ąes escal├Īveis com o suporte de ferramentas externas como caches e balanceadores de carga.GIS em 3 horasMiguel Galves
╠²
O documento fornece uma introdu├¦├Żo sobre Sistemas de Informa├¦├Żo Geogr├Īfica (SIG), explicando conceitos b├Īsicos como georreferenciamento, modelos vetoriais e raster, proje├¦├Ąes cartogr├Īficas, armazenamento de dados espaciais e protocolos de acesso como WMS e WFS.AJAXMiguel Galves
╠²
O documento apresenta um hist├│rico da web, como evoluiu a arquitetura de sites e a introdu├¦├Żo do AJAX. Explica que AJAX utiliza tecnologias j├Ī existentes como JavaScript, XML e HTTP para permitir atualiza├¦├Ąes ass├Łncronas na p├Īgina e maior interatividade. Tamb├®m destaca os desafios do desenvolvimento com AJAX, como compatibilidade entre navegadores e a necessidade de implementar mecanismos como hist├│rico de navega├¦├Żo.Data Mining em redes sociaisMiguel Galves
╠²
O documento descreve experimentos de an├Īlise de dados em redes sociais para determinar not├Łcias importantes, medir opini├Ąes de torcedores em tempo real e verificar o valor de marcas com base no engajamento online. Os experimentos coletam dados do Twitter, Facebook e outras plataformas usando APIs para processar grandes volumes de dados.Ad
Recently uploaded (20)
HOW TO FACE THREATS FROM THE FORCES OF NATURE EXISTING ON PLANET EARTH.pdf
HOW TO FACE THREATS FROM THE FORCES OF NATURE EXISTING ON PLANET EARTH.pdfFaga1939
╠²
This article aims to present how to deal with localized or global threats to human beings caused by the forces of nature that exist on planet Earth. The threats to human beings caused by the forces of nature that exist on planet Earth include earthquakes and tsunamis that can affect specific areas of the planet, volcanic eruptions that can affect specific areas of the planet or have a global impact, the cooling of the Earth's core and the inversion of the Earth's magnetic poles, both of which have a global impact. Specific measures are proposed to deal with each of the threats, but in addition to these, it is necessary to create a global structure, a World Organization for Defense Against Natural Disasters with global scope linked to the UN (United Nations) that has the capacity to technically coordinate the actions of countries around the world in dealing with these threats. The creation of this body is absolutely necessary because most of the threats from the forces of nature that exist on planet Earth have a global impact. To carry out its functions, this body must have financial resources from a global fund against natural disasters of global scope to be maintained by all countries on the planet and administered by the UN.Gas Exchange in Insects and structures 01
Gas Exchange in Insects and structures 01PhoebeAkinyi1
╠²
Description of structures involved in gaseous exchangeMatt Ridley: Economic Evolution and Ideas that have Sex
Matt Ridley: Economic Evolution and Ideas that have SexConservative Institute / Konzervat├Łvny in┼Ītit├║t M. R. ┼Ātef├Īnika
╠²
Matt Ridley is a British independent science popularizer, journalist, entrepreneur, and author of several bestselling books, for which he has received numerous awards. He focuses primarily on the economic, biological, and environmental dimensions of the spontaneous functioning and advancement of human society. GBSN_ Unit 1 - Introduction to Microbiology
GBSN_ Unit 1 - Introduction to MicrobiologyAreesha Ahmad
╠²
Microbiology for Nursing students - According to New PNC course curriculum - 2025Science Holiday Homework (interesting slide )
Science Holiday Homework (interesting slide )aryanxkohli88
╠²
science holiday homework
good for childrenIntroduction to Microbiology and Microscope
Introduction to Microbiology and Microscopevaishrawan1
╠²
This presentation describe about the types of microscope, history and development of microscope and microbiology. Especially focus on Structure & Morphology of BacteriaScience Experiment: Properties of Water.pptx
Science Experiment: Properties of Water.pptxmarionrada1985
╠²
Different experiments to know the properties of waterCULTIVATION - HARVESTING - PROCESSING - STORAGE -.pdf
CULTIVATION - HARVESTING - PROCESSING - STORAGE -.pdfNistarini College, Purulia (W.B) India
╠²
This presentation offers a brief idea about the cultivation, processing, storage and marketing of medicinal plants for the health care practices in sustainable health. The role of different factors has been assessed for the same.Primary and Secondary immune modulation.pptx
Primary and Secondary immune modulation.pptxdevikasanalkumar35
╠²
Primary and secondary immune modulation
Smart Grids Selected Topics, Advanced Metering Infrastructure
Smart Grids Selected Topics, Advanced Metering InfrastructureFrancisSeverineRugan
╠²
This slides are for the smart grid AMI componentAbzymes mimickers in catalytic reactions at nanoscales
Abzymes mimickers in catalytic reactions at nanoscalesOrchideaMariaLecian
╠²
Title: Abzymes mimickers in
catalytic reactions at nanoscales
Speaker: Orchidea Maria Lecian
Authors: Orchidea Maria Lecian, Sergey Suchkov
Talk presented at 4th International Conference on
Advanced Nanomaterials and Nanotechnology
12-13 June 2025, Rome, Italy on 12 June 2025.Matt Ridley: Economic Evolution and Ideas that have Sex
Matt Ridley: Economic Evolution and Ideas that have SexConservative Institute / Konzervat├Łvny in┼Ītit├║t M. R. ┼Ātef├Īnika
╠²
Ad
Comparison of Genomic DNA to cDNA Alignment Methods
- 1. Comparison of Genomic DNA to cDNA Alignment Methods Miguel Galves and Zanoni Dias Institute of Computing ŌĆō Unicamp ŌĆō Campinas ŌĆō SP ŌĆō Brazil {miguel.galves,zanoni}@ic.unicamp.br Scylla Bioinformatics ŌĆō Campinas ŌĆō SP ŌĆō Brazil {miguel,zanoni}@scylla.com.br
- 2. Agenda ’ü¼ Introduction ’ü¼ Problem ’ü¼ Aligners ’ü¼ Data set ’ü¼ Subsets ’ü¼ Evaluation Methods ’ü¼ Results: Exact Alignments ’ü¼ Results: EST Alignments ’ü¼ Running Time Comparison ’ü¼ Conclusions
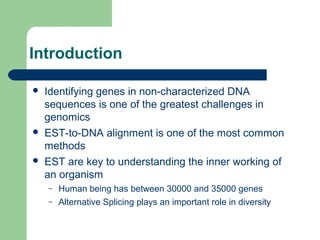
- 3. Introduction ’ü¼ Identifying genes in non-characterized DNA sequences is one of the greatest challenges in genomics ’ü¼ EST-to-DNA alignment is one of the most common methods ’ü¼ EST are key to understanding the inner working of an organism ŌĆō Human being has between 30000 and 35000 genes ŌĆō Alternative Splicing plays an important role in diversity
- 4. CCCGGGAAACGAAUAU CCUCUCACCCGGGA CUUGGCCCGGGAAACGAAUAU CCUCUCACCCGGG A CUUGG Problem Mature mRNA mRNA Intron Exon
- 5. Problem: How to solve ? ’ü¼ Classic algorithms ŌĆō Dynamic programming ’ü¼ Heuristic based algorithms ŌĆō Multi-steps ŌĆō Based on other tools such as Blast and local alignments.
- 6. Aligners ’ü¼ Java version of global and semi-global ŌĆō Affine gap penalty function ŌĆō Linear space ŌĆō Global algorithm by Miller and Myers (1988) ŌĆō Semi-global based on global algorithm ’ü¼ Heuristic based algorithms ŌĆō sim4, Spidey and est_genome
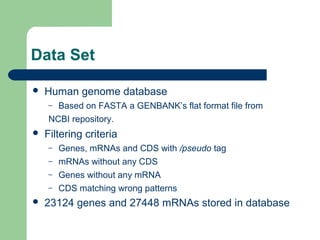
- 7. Data Set ’ü¼ Human genome database ŌĆō Based on FASTA a GENBANKŌĆÖs flat format file from NCBI repository. ’ü¼ Filtering criteria ŌĆō Genes, mRNAs and CDS with /pseudo tag ŌĆō mRNAs without any CDS ŌĆō Genes without any mRNA ŌĆō CDS matching wrong patterns ’ü¼ 23124 genes and 27448 mRNAs stored in database
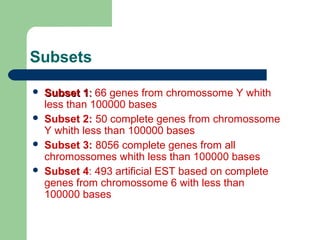
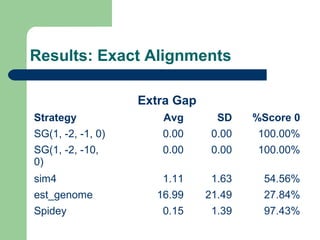
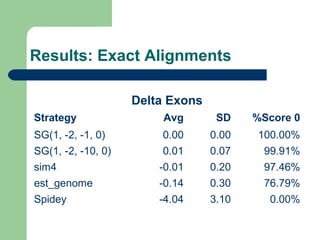
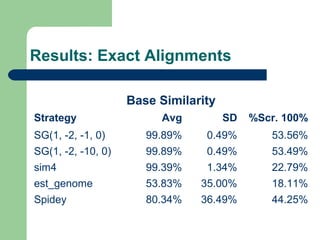
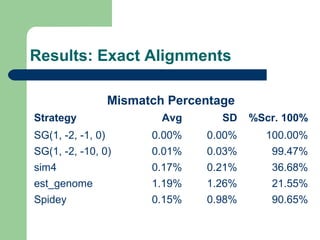
- 8. Subsets ’ü¼ Subset 1Subset 1:: 66 genes from chromossome Y whith less than 100000 bases ’ü¼ Subset 2: 50 complete genes from chromossome Y whith less than 100000 bases ’ü¼ Subset 3: 8056 complete genes from all chromossomes whith less than 100000 bases ’ü¼ Subset 4: 493 artificial EST based on complete genes from chromossome 6 with less than 100000 bases
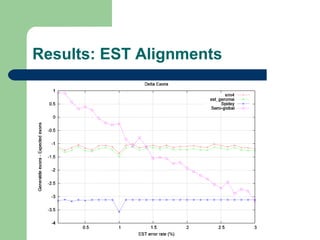
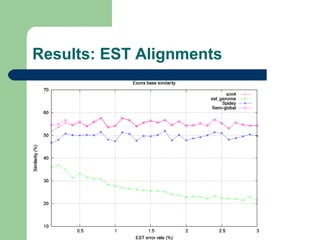
- 9. Evaluation methods ’ü¼ Number of gaps introduced in the aligned gene sequence ’ü¼ Delta exons ’ü¼ Bases similarity percentage ’ü¼ Mismatch percentage
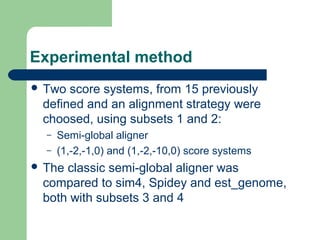
- 10. Experimental method ’ü¼ Two score systems, from 15 previously defined and an alignment strategy were choosed, using subsets 1 and 2: ŌĆō Semi-global aligner ŌĆō (1,-2,-1,0) and (1,-2,-10,0) score systems ’ü¼ The classic semi-global aligner was compared to sim4, Spidey and est_genome, both with subsets 3 and 4
- 11. Results: Exact Alignments Extra Gap Strategy Avg SD %Score 0 SG(1, -2, -1, 0) 0.00 0.00 100.00% SG(1, -2, -10, 0) 0.00 0.00 100.00% sim4 1.11 1.63 54.56% est_genome 16.99 21.49 27.84% Spidey 0.15 1.39 97.43%
- 12. Results: Exact Alignments Delta Exons Strategy Avg SD %Score 0 SG(1, -2, -1, 0) 0.00 0.00 100.00% SG(1, -2, -10, 0) 0.01 0.07 99.91% sim4 -0.01 0.20 97.46% est_genome -0.14 0.30 76.79% Spidey -4.04 3.10 0.00%
- 13. Results: Exact Alignments Base Similarity Strategy Avg SD %Scr. 100% SG(1, -2, -1, 0) 99.89% 0.49% 53.56% SG(1, -2, -10, 0) 99.89% 0.49% 53.49% sim4 99.39% 1.34% 22.79% est_genome 53.83% 35.00% 18.11% Spidey 80.34% 36.49% 44.25%
- 14. Results: Exact Alignments Mismatch Percentage Strategy Avg SD %Scr. 100% SG(1, -2, -1, 0) 0.00% 0.00% 100.00% SG(1, -2, -10, 0) 0.01% 0.03% 99.47% sim4 0.17% 0.21% 36.68% est_genome 1.19% 1.26% 21.55% Spidey 0.15% 0.98% 90.65%
- 17. Running Time Comparison EST-to-DNA (sec/alignment) mRNA-toDNA (sec/alignment) sim4 0.013 0.170 Spidey 0.066 0.140 est_genome 0.640 3.400 Semi-global 0.670 5.170
- 18. Conclusions ’ü¼ Classic semi-globl algorithm produces good results ŌĆō Running time is a problem, although it can be improved ’ü¼ Sim4 produces the best results amont external softwares tested
- 19. Thanks
