ChemEngine ACS
- 1. ChemEngine: Harvest Chemical Data from PDF Files M. Karthikeyan, Ph.D CSIR-National Chemical Laboratory http://moltable.ncl.res.in/ Pune, India
- 2. ChemEngine
- 4. ChemEngine • Digital access to chemical journals resulted in a vast array of molecular information that is now available in the supplementary material files in PDF format. However, extracting this molecular information, generally from a PDF document format is a daunting task. • Here we present an approach to harvest 3D molecular data from the supporting information of scientific research articles that are normally available from publisher’s resources. • In order to demonstrate the feasibility of extracting truly computable molecules from PDF file formats in a fast and efficient manner, we have developed an application, namely ChemEngine. • This program recognizes textual patterns from the supplementary data and generates standard molecular structure data (bond matrix, atomic coordinates) that can be subjected to a multitude of computational processes automatically.
- 5. ChemEngine • The methodology has been demonstrated via three case studies on different formats of coordinates data stored in supplementary information files, wherein ChemEngine selectively harvested the atomic coordinates and interpreted them as molecules with high accuracy. • The reusability of extracted molecular coordinate data was demonstrated by computing Single Point Energies (SPEs) that were in close agreement with the original computed data provided with the articles. • It is envisaged that the methodology will enable large scale conversion of molecular information from supplementary files available in PDF format into a collection of ready- to- compute molecular data to create an automated workflow for advanced computational processes. • Software in the form of jar file is available for downloading at the Sourceforge site.
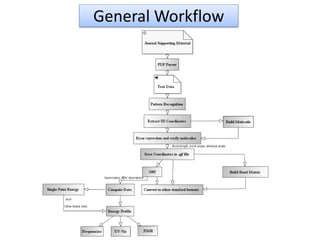
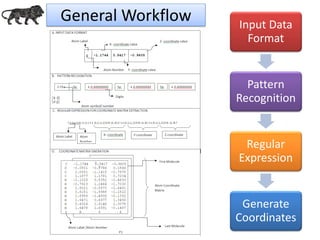
- 7. General Workflow Input Data Format Pattern Recognition Regular Expression Generate Coordinates
- 8. Pseudo code: (Co-ordinate Text).matches(“Regular Expression Pattern with Delimiter Definition”); For Example: Delimiter: Comma String_Data.matches("^[A-Za-z0- 9]{1,2},[0]{0,1}[,]{0,1}-{0,1}.{1,2}[0-9]{1,10},- {0,1}.{1,2}[0-9]{1,10}.{1,}") Delimiter: Space String_Data.matches("^[A-Za-z0- 9]{1,2}s+[0]{0,1}[s+]{0,1}-{0,1}.{1,2}[0-9]{0,10}s+- {0,1}.{1,2}[0-9]{0,10}.{1,}") Pattern Recognition
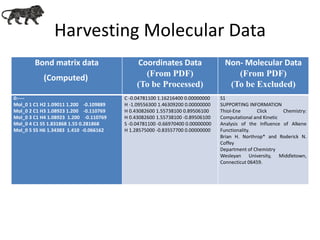
- 9. Harvesting Molecular Data Bond matrix data (Computed) Coordinates Data (From PDF) (To be Processed) Non- Molecular Data (From PDF) (To be Excluded) 0:---- Mol_0 1 C1 H2 1.09011 1.200 -0.109889 Mol_0 2 C1 H3 1.08923 1.200 -0.110769 Mol_0 3 C1 H4 1.08923 1.200 -0.110769 Mol_0 4 C1 S5 1.831868 1.55 0.281868 Mol_0 5 S5 H6 1.34383 1.410 -0.066162 C -0.04781100 1.16216400 0.00000000 H -1.09556300 1.46309200 0.00000000 H 0.43082600 1.55738100 0.89506100 H 0.43082600 1.55738100 -0.89506100 S -0.04781100 -0.66970400 0.00000000 H 1.28575000 -0.83557700 0.00000000 S1 SUPPORTING INFORMATION Thiol-Ene Click Chemistry: Computational and Kinetic Analysis of the Influence of Alkene Functionality. Brian H. Northrop* and Roderick N. Coffey Department of Chemistry Wesleyan University, Middletown, Connecticut 06459.
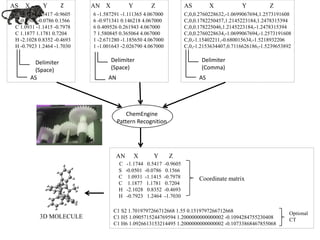
- 10. AS C -1.1744 0.5417 -0.9605 S -0.0501 -0.0786 0.1566 C 1.0931 -1.1415 -0.7978 C 1.1877 1.1781 0.7204 H -2.1028 0.8352 -0.4693 H -0.7923 1.2464 -1.7030 6 -1.587291 -1.111365 4.067000 6 -0.971341 0.146218 4.067000 6 0.409526 0.261943 4.067000 7 1.580845 0.365064 4.067000 1 -2.671280 -1.185650 4.067000 1 -1.001643 -2.026790 4.067000 AS Delimiter (Space) AN Delimiter (Space) Delimiter (Comma) C,0,0.2760228632,-1.0699067694,1.2573191608 C,0,0.1782250457,1.2145223184,1.2478315394 C,0,0.178225046,1.2145223184,-1.2478315394 C,0,0.2760228634,-1.0699067694,-1.2573191608 C,0,-1.15402211,-0.680015634,-1.5218932206 C,0,-1.2153634407,0.7116626186,-1.5239653892 ChemEngine Pattern Recognition C -1.1744 0.5417 -0.9605 S -0.0501 -0.0786 0.1566 C 1.0931 -1.1415 -0.7978 C 1.1877 1.1781 0.7204 H -2.1028 0.8352 -0.4693 H -0.7923 1.2464 -1.7030 C1 S2 1.7019797266712668 1.55 0.1519797266712668 C1 H5 1.0905715244769594 1.2000000000000002 -0.1094284755230408 C1 H6 1.0926613153214495 1.2000000000000002 -0.10733868467855068 X Y ZAS X Y ZAN AS X Y Z AN X Y Z Optional CT Coordinate matrix 3D MOLECULE
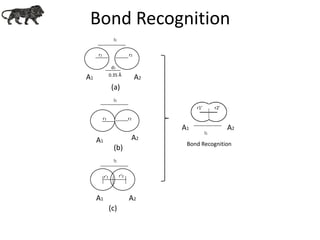
- 11. Bond Recognition A1 A2 r1 r2 l1 l1 Bond Recognition A1 A2 l1 r1’ r2’ l1 A1 A2 r1 r2 d1 0.35 Å A1 A2 r'1 r‘2 (a) (b) (c)
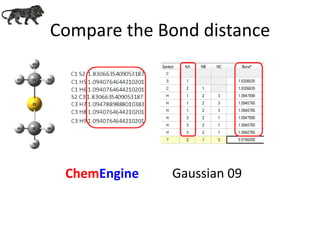
- 12. Compare the Bond distance ChemEngine Gaussian 09
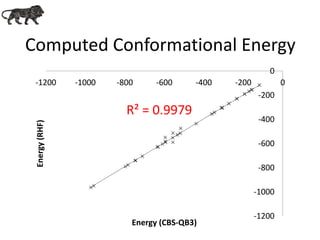
- 13. Computed Conformational Energy R² = 0.9979 -1200 -1000 -800 -600 -400 -200 0 -1200 -1000 -800 -600 -400 -200 0 Energy(RHF) Energy (CBS-QB3)
- 14. Case Studies Entry Case Study N= molecules Regular Expression pattern Format & Delimit er 1 Epoxide formation from sulfur ylides and aldehydes 29 ^[A-Za-z0-9]{1,2}s+- {0,1}.{1,2}[0-9]{1,8}s+- {0,1}.{1,2}[0-9]{1,8}.{1,} PDF Space 2 Thiol ene click chemistry 115 ^[A-Za-z0-9]{1,2}s+- {0,1}.{1,2}[0-9]{1,8}s+- {0,1}.{1,2}[0-9]{1,8}.{1,} Text Space 3 Design of tetra(arenediyl)bis(allyl) derivatives for cope rearrangement transition states 55 ^[A-Za-z0- 9]{1,2},[0]{0,1}[,]{0,1}- {0,1}.{1,2}[0-9]{1,10},- {0,1}.{1,2}[0-9]{1,10}.{1,} PDF Comma
- 15. ChemEngine
- 17. Thank You



![Pseudo code:
(Co-ordinate Text).matches(“Regular Expression Pattern
with Delimiter Definition”);
For Example: Delimiter: Comma
String_Data.matches("^[A-Za-z0-
9]{1,2},[0]{0,1}[,]{0,1}-{0,1}.{1,2}[0-9]{1,10},-
{0,1}.{1,2}[0-9]{1,10}.{1,}")
Delimiter: Space
String_Data.matches("^[A-Za-z0-
9]{1,2}s+[0]{0,1}[s+]{0,1}-{0,1}.{1,2}[0-9]{0,10}s+-
{0,1}.{1,2}[0-9]{0,10}.{1,}")
Pattern Recognition](https://image.slidesharecdn.com/6ad32f07-a8fc-4f2c-a487-15acf6122c8e-160401155107/85/ChemEngine-ACS-8-320.jpg)
![Case Studies
Entry Case Study N= molecules Regular Expression pattern Format
&
Delimit
er
1 Epoxide formation
from sulfur ylides and
aldehydes
29 ^[A-Za-z0-9]{1,2}s+-
{0,1}.{1,2}[0-9]{1,8}s+-
{0,1}.{1,2}[0-9]{1,8}.{1,}
PDF
Space
2 Thiol ene click chemistry 115 ^[A-Za-z0-9]{1,2}s+-
{0,1}.{1,2}[0-9]{1,8}s+-
{0,1}.{1,2}[0-9]{1,8}.{1,}
Text
Space
3 Design of
tetra(arenediyl)bis(allyl)
derivatives for cope
rearrangement
transition states
55 ^[A-Za-z0-
9]{1,2},[0]{0,1}[,]{0,1}-
{0,1}.{1,2}[0-9]{1,10},-
{0,1}.{1,2}[0-9]{1,10}.{1,}
PDF
Comma](https://image.slidesharecdn.com/6ad32f07-a8fc-4f2c-a487-15acf6122c8e-160401155107/85/ChemEngine-ACS-14-320.jpg)