p3d @EuroSciPy2010 by C. Fufezan
- 1. High-throughput structural bioinformatics using Python & p3d Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 2. Overview Background p3d overview example ATP binding site Fufezan, C. and Specht M. (2009) BMC Bioinformatics 10, 258 http://p3d.fufezan.net http://github.com/fu/p3d Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) clone us - fork us!
- 3. Background Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
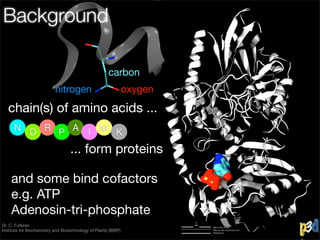
- 4. Background carbon nitrogen oxygen chain(s) of amino acids ... N D R P A I M K ... form proteins Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 5. Background carbon nitrogen oxygen chain(s) of amino acids ... N D R P A I M K ... form proteins Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 6. Background carbon nitrogen oxygen chain(s) of amino acids ... N D R P A I M K ... form proteins and some bind cofactors e.g. ATP Adenosin-tri-phosphate Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 7. Background knowledge based approaches to elucidate structural factors that are essential for co-factor binding - protein engineering proteins - protein folding - co-factor tuning Proteins_c1_sp_Ob.qxp 9/11/08 4:14 PM Page 1 proteins STRUCTURE O FUNCTION O BIOINFORMATICS V O LU M E 7 3 , N U M B E R 3 , N OV E M B E R 1 5 , V O LU M E 7 3 , N U M B E R 3 , N OV E M B E R 1 5 , 2 0 0 8 PAG E S Morozov et al. (2004) PNAS, 101, 6946Huang et al.(2004) PNAS, 101, 5536Fufezan et al. (2008) Proteins, 73, 690Negron et al. (2009) Proteins, 74, 400Fufezan (2010) Proteins, in press 5 2 7 ¨C 7 9 4 Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) A PDB Survey of Heme Ligands in Proteins 2 0 0 8
- 8. p3d Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 9. p3d overview Fufezan, C. and Specht M. (2009) BMC Bioinformatics 10, 258 http://p3d.fufezan.net Python module that allows to access and manipulate protein structure ?les rapid development of new screening tools easily incorporate complex queries Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 10. chain atom AA resid type x y z user idx ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) beta or temperature factor O C N CA CB CG1 CG2
- 11. Protein Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) protein ... not-proteinogenic hash chain['A'] ... oxygen nitrogen backbone atype['CA'] ... oxygen backbone alpha residues protein not protein
- 12. Protein Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) protein ... not-proteinogenic hash chain['A'] ... oxygen nitrogen backbone atype['CA'] ... oxygen backbone alpha residues protein not protein
- 13. Protein Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) protein ... not-proteinogenic hash chain['A'] ... oxygen nitrogen backbone atype['CA'] ... oxygen backbone alpha residues protein not protein
- 14. Protein Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) protein ... not-proteinogenic hash chain['A'] ... oxygen nitrogen backbone atype['CA'] ... oxygen backbone alpha residues protein not protein
- 15. Protein Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) protein ... not-proteinogenic hash chain['A'] ... oxygen nitrogen backbone atype['CA'] ... oxygen backbone alpha residues protein not protein
- 16. Tree Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 17. Tree Object ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 18. Tree Object query( Vector1, radius ) ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 19. Tree Object query( Vector1, radius ) ATOM Object idx atype aa chain x y z user beta protein-object Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) Vectors do not have to be atoms!!
- 20. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone alpha residues protein not protein
- 21. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone alpha residues protein not protein Query function using human readable syntax e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯)
- 22. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) residues protein not protein
- 23. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) residues protein not protein
- 24. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) pdb.query(¡®oxygen and not protein¡¯) residues protein not protein
- 25. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) pdb.query(¡®oxygen and not protein¡¯) residues pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) ) protein not protein
- 26. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) pdb.query(¡®oxygen and not protein¡¯) residues pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) ) protein not protein for residueName in pdb.hash[non-aa-resname]:
- 27. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) pdb.query(¡®oxygen and not protein¡¯) residues pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) ) protein not protein for residueName in pdb.hash[non-aa-resname]: targets = pdb.query(' protein and within 4 of
- 28. Protein class List of atom objects (vectors) sets (hashes) oxygen BSP Tree O1 FME A 1 CA PHE A 2 backbone Query function using human readable syntax alpha e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯) pdb.query(¡®resname HOH and within 4 of resname ASP¡¯) pdb.query(¡®oxygen and not protein¡¯) residues pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) ) protein not protein for residueName in pdb.hash[non-aa-resname]: targets = pdb.query(' protein and within 4 of ( resname 'residueName' and oxygen )' )
- 29. Example ATP binding Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 30. The ATP binding sites Adenosine-tri-phosphate ¦¤G?' = -30 kJ mol-1 40 kg / day Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 31. The ATP binding sites non. redundant set of proteins 24 binding sites Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 32. The ATP binding sites non. redundant set of proteins 24 binding sites Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 33. The ATP binding sites non. redundant set of proteins 24 binding sites Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 34. The ATP binding sites non. redundant set of proteins 24 binding sites Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 35. Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 36. The ATP binding site Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 37. The ATP binding site Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 38. The ATP binding site +4.5 hydropathy index Observations 10 0 1 -4.5 Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP) non. redundant set of proteins 24 binding sites
- 39. Summary p3d allows to develop quickly Python scripts to screen Protein structures combines Vectors, sets and BSPTree p3d allows ?exible and complex queries using human readable language Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)
- 40. Acknowledgements M. Specht Prof. Dr. M. Hippler founding by the DFG and Alexander von Humboldt Stiftung Dr. C. Fufezan Institute for Biochemistry and Biotechnology of Plants (IBBP)









![Protein Object
ATOM Object
idx
atype
aa
chain
x
y
z
user
beta
protein-object
Dr. C. Fufezan
Institute for Biochemistry and Biotechnology of Plants (IBBP)
protein
... not-proteinogenic
hash
chain['A']
...
oxygen
nitrogen
backbone
atype['CA']
...
oxygen
backbone
alpha
residues
protein
not protein](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-11-320.jpg)
![Protein Object
ATOM Object
idx
atype
aa
chain
x
y
z
user
beta
protein-object
Dr. C. Fufezan
Institute for Biochemistry and Biotechnology of Plants (IBBP)
protein
... not-proteinogenic
hash
chain['A']
...
oxygen
nitrogen
backbone
atype['CA']
...
oxygen
backbone
alpha
residues
protein
not protein](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-12-320.jpg)
![Protein Object
ATOM Object
idx
atype
aa
chain
x
y
z
user
beta
protein-object
Dr. C. Fufezan
Institute for Biochemistry and Biotechnology of Plants (IBBP)
protein
... not-proteinogenic
hash
chain['A']
...
oxygen
nitrogen
backbone
atype['CA']
...
oxygen
backbone
alpha
residues
protein
not protein](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-13-320.jpg)
![Protein Object
ATOM Object
idx
atype
aa
chain
x
y
z
user
beta
protein-object
Dr. C. Fufezan
Institute for Biochemistry and Biotechnology of Plants (IBBP)
protein
... not-proteinogenic
hash
chain['A']
...
oxygen
nitrogen
backbone
atype['CA']
...
oxygen
backbone
alpha
residues
protein
not protein](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-14-320.jpg)
![Protein Object
ATOM Object
idx
atype
aa
chain
x
y
z
user
beta
protein-object
Dr. C. Fufezan
Institute for Biochemistry and Biotechnology of Plants (IBBP)
protein
... not-proteinogenic
hash
chain['A']
...
oxygen
nitrogen
backbone
atype['CA']
...
oxygen
backbone
alpha
residues
protein
not protein](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-15-320.jpg)










![Protein class
List of atom
objects (vectors)
sets (hashes)
oxygen
BSP Tree
O1
FME A
1
CA
PHE A
2
backbone
Query function using human readable syntax
alpha
e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯)
pdb.query(¡®resname HOH and within 4 of resname ASP¡¯)
pdb.query(¡®oxygen and not protein¡¯)
residues
pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) )
protein
not protein
for residueName in pdb.hash[non-aa-resname]:](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-26-320.jpg)
![Protein class
List of atom
objects (vectors)
sets (hashes)
oxygen
BSP Tree
O1
FME A
1
CA
PHE A
2
backbone
Query function using human readable syntax
alpha
e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯)
pdb.query(¡®resname HOH and within 4 of resname ASP¡¯)
pdb.query(¡®oxygen and not protein¡¯)
residues
pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) )
protein
not protein
for residueName in pdb.hash[non-aa-resname]:
targets = pdb.query(' protein and within 4 of](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-27-320.jpg)
![Protein class
List of atom
objects (vectors)
sets (hashes)
oxygen
BSP Tree
O1
FME A
1
CA
PHE A
2
backbone
Query function using human readable syntax
alpha
e.g.: pdb.query(¡®backbone and resid 5..12 and within 5 of resname FME¡¯)
pdb.query(¡®resname HOH and within 4 of resname ASP¡¯)
pdb.query(¡®oxygen and not protein¡¯)
residues
pdb.query(¡® protein and within 4 of ¡¯, p3d.vector.Vector(x,y,z) )
protein
not protein
for residueName in pdb.hash[non-aa-resname]:
targets = pdb.query(' protein and within 4 of
( resname 'residueName' and oxygen )' )](https://image.slidesharecdn.com/cfeuroscipy2010-131212100524-phpapp02/85/p3d-EuroSciPy2010-by-C-Fufezan-28-320.jpg)











