Comparison Of Commercially Available Str Typing Kits (Nx Power Lite)
- 1. COMPARISON OF COMMERCIALLY AVAILABLE STR TYPING KITS: SAMPLE CONDITION, ACCURACY, AND EFFICACY By Courtney Brennan Special thanks to my Mentors/Professors: M. Guido, K. Sweder, and M. Sponsler
- 2. BACKGROUND  Many labs use commercially available “kits” to type DNA samples  These are usually “STR” (short tandem repeat) typing kits.  Many labs only use one kit, and most forensic labs only use one kit per sample.  The two most common producers of these kits are Applied Biosystems and Promega Corporation.  Many kits include the 13 core loci (CODIS)  A Loci is a specific location on a chromosome where a sequence or gene is found  Variants of this sequence are known as alleles
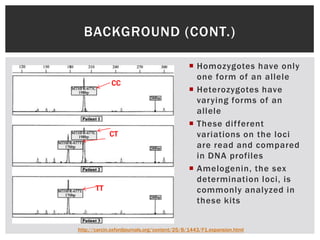
- 3. BACKGROUND (CONT.)  Homozygotes have only one form of an allele CC  Heterozygotes have varying forms of an allele  These dif ferent CT variations on the loci are read and compared in DNA profiles  Amelogenin, the sex determination loci, is TT commonly analyzed in these kits http://carcin.oxfordjournals.org/content/25/8/1443/F1.expansion.html
- 4. UNDERSTANDING ACCURACY While kits tend to be far within the acceptable range of accuracy for typing, accuracy has costs Increased accuracy may decrease the real-life application of the kit since contaminants or mixed profiles would be more difficult to analyze Some samples with only a small amount of DNA can exhibit inaccurate readings if they have low copy number
- 5. ANALYSIS OF AMPFISTR® SGM PLUS™ MULTIPLEX SYSTEM FOR EFFECTIVENESS ON LOW COPY NUMBER SAMPLES AMPFISTR® SGM Plus™ system was tested for usefulness in analyzing LCN (low copy number) samples Low copy number generally refers to samples in which there is less than 100pg of input genetic material This approach can have issues – sometimes one allele is not present in a large enough amount to be detected properly
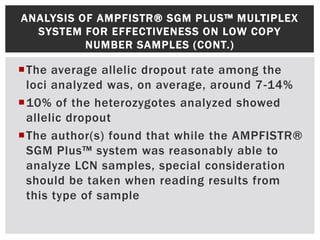
- 6. ALLELIC DROPOUT IN LCN CONDITIONS Difference # Allelic dropout (% of Locus observed in 28 vs. Alleles expected) 34 cycles Amelogenin 6 [4H] 4 (14.8) D3S1358 8 [3L, 2H] 5 (10.6) HumVWF31 10 [5L, 2H] 7 (10.4) D16S539 10 [4L, 4H] 8 (17) D2S1338 13 [4L, 2H] 6 (8.9) D8S1179 11 [2L, 2H] 6( 8.9) D21S11 6 [4L, 2H] 4 (14.8) D18S51 11 [4L, 2H] 6( 8.9) D19S433 6 [4L, 2H] 6( 8.9) HUMTHO1 1 [1H] 1 (3.7) HumFIBRA 13 [2L, 4H] 5 (7.4) • Allelic dropout is when one or more of the alleles present in a sample are either poorly detected or undetected in a profile • This results in false homozygosity – being led to believe a specific loci is homozygous when in fact the analysis simply failed to pick up the second allele
- 7. ANALYSIS OF AMPFISTR® SGM PLUS™ MULTIPLEX SYSTEM FOR EFFECTIVENESS ON LOW COPY NUMBER SAMPLES (CONT.) The average allelic dropout rate among the loci analyzed was, on average, around 7-14% 10% of the heterozygotes analyzed showed allelic dropout The author(s) found that while the AMPFISTR® SGM Plus™ system was reasonably able to analyze LCN samples, special consideration should be taken when reading results from this type of sample
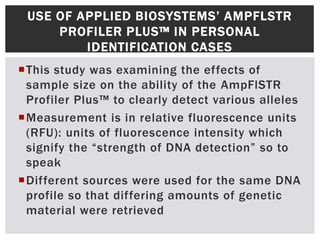
- 8. USE OF APPLIED BIOSYSTEMS’ AMPFLSTR PROFILER PLUS™ IN PERSONAL IDENTIFICATION CASES This study was examining the effects of sample size on the ability of the AmpFlSTR Profiler Plus™ to clearly detect various alleles Measurement is in relative fluorescence units (RFU): units of fluorescence intensity which signify the “strength of DNA detection” so to speak Different sources were used for the same DNA profile so that differing amounts of genetic material were retrieved
- 9. DNA AMOUNTS AS MEASURED BY RFU Heterozygous DNA (ng) D3S1358[L/H] VWA[L/H] FGA[L/H] D18S51[L/H] D7S820[L/H] 4 1291/1181 1119/907 701/678 798/601 591/459 2 1005/927 773/693 593/538 633/519 486/390 1 655/695 664/435 499/441 597/575 340/407 0.5 385/341 225/212 229/218 197/226 173/155 0.25 132/169 127/101 104/106 114/66 63/77 0.125 78/107 75/68 56/59 55/39 41/45 0.063 51/42 45/47 37/38 38/31 28/31 0.031 47/41 - - 28/26 - Homozygous DNA (ng) X D8S1179 D21S11 D5S818 D13S317 4 2619 2221 2018 1789 1162 2 1608 1624 1309 1368 1013 1 1131 906 1000 1110 1004 0.5 738 446 440 484 431 0.25 343 262 177 244 204 0.125 180 136 122 112 108 0.063 95 71 61 62 58 0.031 40 29 47 25 35
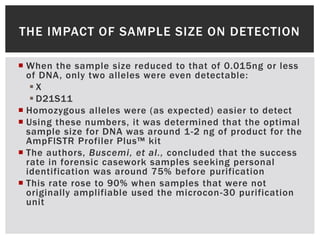
- 10. THE IMPACT OF SAMPLE SIZE ON DETECTION  When the sample size reduced to that of 0.015ng or less of DNA, only two alleles were even detectable: X  D21S11  Homozygous alleles were (as expected) easier to detect  Using these numbers, it was determined that the optimal sample size for DNA was around 1-2 ng of product for the AmpFlSTR Profiler Plus™ kit  The authors, Buscemi, et al., concluded that the success rate in forensic casework samples seeking personal identification was around 75% before purification  This rate rose to 90% when samples that were not originally amplifiable used the microcon -30 purification unit
- 11. AMPFLSTR® IDENTIFILER® AND MOTHERLESS PATERNITY TESTS  A study was done to determine the effectiveness of the AmpFLSTR® Identifiler® kit in cases where the mother’s sample was not available for comparison in paternity testing  Paternity testing generally involves three samples; this method was referred to as the “trio” of samples:  Potential father  Mother  Child  In some cases, the mother’s sample was not available and testing was only done between potential father and child. This was referred to as “duo” testing.
- 12. VALIDIT Y OF MOTHERLESS PATERNIT Y TESTING  It was concluded that the results from duo, rather than trio, testing using the AmpFLSTR® Identifiler® kit were not sufficient to prove paternity:  In trio cases, the mean value of probability of paternity was 99.999997%, with a minimum of four excluding loci  In duo cases, over half of the cases were unable to reach the accepted (99.999%) probability of paternity value.  This is logical due to the fact that in duo cases, sometimes as few as 1 excluding loci were found  Consider: Do other kits have similar results when testing in this manner?
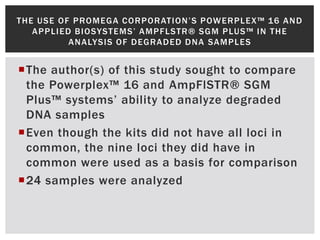
- 13. T HE USE OF P ROM E G A C ORPORAT I ON’S POW E RPLE X ™ 16 A N D A P P LI E D BI OSYST E MS’ A M P F LSTR® SG M P LUS™ I N T H E A N ALYSIS OF DE G RA DE D DN A SA M P LE S The author(s) of this study sought to compare the Powerplex™ 16 and AmpFlSTR® SGM Plus™ systems’ ability to analyze degraded DNA samples Even though the kits did not have all loci in common, the nine loci they did have in common were used as a basis for comparison 24 samples were analyzed
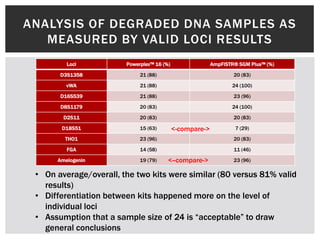
- 14. ANALYSIS OF DEGRADED DNA SAMPLES AS MEASURED BY VALID LOCI RESULTS Loci Powerplex™ 16 (%) AmpFlSTR® SGM Plus™ (%) D3S1358 21 (88) 20 (83) vWA 21 (88) 24 (100) D16S539 21 (88) 23 (96) D8S1179 20 (83) 24 (100) D2S11 20 (83) 20 (83) D18S51 15 (63) <-compare-> 7 (29) THO1 23 (96) 20 (83) FGA 14 (58) 11 (46) Amelogenin 19 (79) <--compare-> 23 (96) • On average/overall, the two kits were similar (80 versus 81% valid results) • Differentiation between kits happened more on the level of individual loci • Assumption that a sample size of 24 is “acceptable” to draw general conclusions
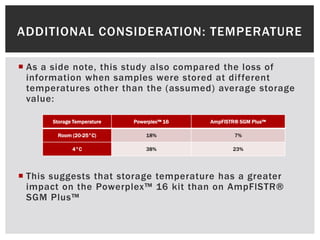
- 15. ADDITIONAL CONSIDERATION: TEMPERATURE  As a side note, this study also compared the loss of information when samples were stored at different temperatures other than the (assumed) average storage value: Storage Temperature Powerplex™ 16 AmpFlSTR® SGM Plus™ Room (20-25°C) 18% 7% 4°C 38% 23%  This suggests that storage temperature has a greater impact on the Powerplex™ 16 kit than on AmpFlSTR® SGM Plus™
- 16. GENOT YPING INCONSISTENCIES - AMPFLSTR® IDENTIFILER® VS. POWERPLEX® 16 13 STRs were used to compare since they were shared between both kits Because different kits use different primers, it was believed that certain inconsistencies might exist between them This issue can be important due to the common Forensic practice of “databasing” Some kits that are produced by the same company can still have primers that are designed differently
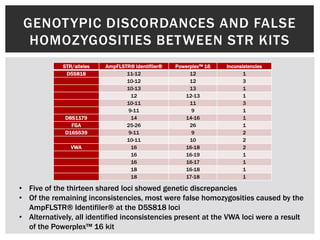
- 17. GENOT YPIC DISCORDANCES AND FALSE HOMOZYGOSITIES BETWEEN STR KITS STR/alleles AmpFLSTR® Identifiler® Powerplex™ 16 Inconsistencies D5S818 11-12 12 1 10-12 12 3 10-13 13 1 12 12-13 1 10-11 11 3 9-11 9 1 D8S1179 14 14-16 1 FGA 25-26 26 1 D16S539 9-11 9 2 10-11 10 2 VWA 16 16-18 2 16 16-19 1 16 16-17 1 18 16-18 1 18 17-18 1 • Five of the thirteen shared loci showed genetic discrepancies • Of the remaining inconsistencies, most were false homozygosities caused by the AmpFLSTR® Identifiler® at the D5S818 loci • Alternatively, all identified inconsistencies present at the VWA loci were a result of the Powerplex™ 16 kit
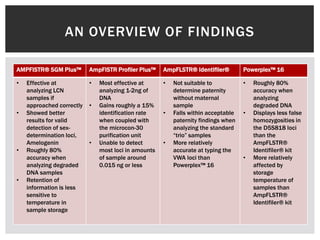
- 18. AN OVERVIEW OF FINDINGS AMPFISTR® SGM Plus™ AmpFlSTR Profiler Plus™ AmpFLSTR® Identifiler® Powerplex™ 16 • Effective at • Most effective at • Not suitable to • Roughly 80% analyzing LCN analyzing 1-2ng of determine paternity accuracy when samples if DNA without maternal analyzing approached correctly • Gains roughly a 15% sample degraded DNA • Showed better identification rate • Falls within acceptable • Displays less false results for valid when coupled with paternity findings when homozygosities in detection of sex- the microcon-30 analyzing the standard the D5S818 loci determination loci, purification unit “trio” samples than the Amelogenin • Unable to detect • More relatively AmpFLSTR® • Roughly 80% most loci in amounts accurate at typing the Identifiler® kit accuracy when of sample around VWA loci than • More relatively analyzing degraded 0.015 ng or less Powerplex™ 16 affected by DNA samples storage • Retention of temperature of information is less samples than sensitive to AmpFLSTR® temperature in Identifiler® kit sample storage
- 19. REFERENCES 1. Butler, John M. Forensic DNA Typing. 2005. Elsevier Academic Press. 2. J. P. Whitaker, E. A. Cotton, P. Gill, “A comparison of the characteristics of profiles produced with the AMPFlSTR(R) SGM Plus(TM) multiplex system for both standard and low copy number (LCN) STR DNA analysis”, Forensic Science International, Volume 123, Issues 2 -3, 1 December 2001, Pages 215 -223, ISSN 0379-0738, DOI: 10.1016/S0379-0738(01)00557-6.(http://www.sciencedirect.com/science/article/B6T6W -44HSMMP- N/2/0a3ac8657953802f439cd3ca16687e38 ) 3. L. Buscemi, M. Pesaresi, C. Sassaroli, F. Alessandrini, A. Tagliabracci, “Further study on suitability of Profiler Plus in personal identification”, International Congress Series, Volume 1239, Progress in Forensic Genetics 9. Proceedings from the 19th, January 2003, Pages 891 -894, ISSN 0531-5131, DOI: 10.1016/S0531- 5131(02)00575-7. (http://www.sciencedirect.com/science/article/B7581 -47W664D- 1NM/2/f01020ea1b32111cf0a42e42d8445492 ) 4. K. Babol-Pokora, R. Jacewicz, Pepinski, S. Szram, “Identifiler(TM) system as an inadequate tool for judging motherless paternity cases”, International Congress Series, Volume 1288, Progress in Forensic Genetics 11 - Proceedings of the 21st International ISFG Congress held in Ponta Delgada, The Azores, Portugal between 13 and 16 September 2005, April 2006, Pages 462 -464, ISSN 0531-5131, DOI: 10.1016/j.ics.2005.09.102. (http://www.sciencedirect.com/science/article/B7581 -4JSBXBX- 5R/2/7e791e028513dce63232f1a5d4d45460 ) 5. B. Glock, R. B. K. Reisacher, S. O. Rennhofer, D. Troscher, E. M. Dauber, W. R. Mayr, “Evaluation of Powerplex(TM) 16 for typing of degraded DNA samples”, International Congress Series, Volume 1239, Progress in Forensic Genetics 9. Proceedings from the 19th, January 2003, Pages 609 -611, ISSN 0531-5131, DOI: 10.1016/S0531-5131(02)00296-0.(http://www.sciencedirect.com/science/article/B7581 -47W664D- 1KB/2/add81b4ca5fbf1a7906672e2318e8a1f ) 6. A. Amorim, C. Alves, L. Pereira, L. Gusmao, “Genotyping inconsistencies and null alleles using AmpFLSTR(R) Identifiler(R) and Powerplex(R) 16 kits”, International Congress Series, Volume 1261, Progress in Forensic Genetics 10, April 2004, Pages 176 -178, ISSN 0531-5131, DOI: 10.1016/S0531-5131(03)01496-1. (http://www.sciencedirect.com/science/article/B7581 -4C4WDDP- 23/2/e458dc9ac25f660bccd6b2cea233202a)



![ALLELIC DROPOUT IN LCN CONDITIONS
Difference #
Allelic dropout (% of
Locus observed in 28 vs. Alleles
expected)
34 cycles
Amelogenin 6 [4H] 4 (14.8)
D3S1358 8 [3L, 2H] 5 (10.6)
HumVWF31 10 [5L, 2H] 7 (10.4)
D16S539 10 [4L, 4H] 8 (17)
D2S1338 13 [4L, 2H] 6 (8.9)
D8S1179 11 [2L, 2H] 6( 8.9)
D21S11 6 [4L, 2H] 4 (14.8)
D18S51 11 [4L, 2H] 6( 8.9)
D19S433 6 [4L, 2H] 6( 8.9)
HUMTHO1 1 [1H] 1 (3.7)
HumFIBRA 13 [2L, 4H] 5 (7.4)
• Allelic dropout is when one or more of the alleles present in a sample
are either poorly detected or undetected in a profile
• This results in false homozygosity – being led to believe a specific loci
is homozygous when in fact the analysis simply failed to pick up the
second allele](https://image.slidesharecdn.com/comparisonofcommerciallyavailablestrtypingkitsnxpowerlite-13324093356203-phpapp01-120322044250-phpapp01/85/Comparison-Of-Commercially-Available-Str-Typing-Kits-Nx-Power-Lite-6-320.jpg)
![DNA AMOUNTS AS MEASURED BY RFU
Heterozygous
DNA (ng) D3S1358[L/H] VWA[L/H] FGA[L/H] D18S51[L/H] D7S820[L/H]
4 1291/1181 1119/907 701/678 798/601 591/459
2 1005/927 773/693 593/538 633/519 486/390
1 655/695 664/435 499/441 597/575 340/407
0.5 385/341 225/212 229/218 197/226 173/155
0.25 132/169 127/101 104/106 114/66 63/77
0.125 78/107 75/68 56/59 55/39 41/45
0.063 51/42 45/47 37/38 38/31 28/31
0.031 47/41 - - 28/26 -
Homozygous
DNA (ng) X D8S1179 D21S11 D5S818 D13S317
4 2619 2221 2018 1789 1162
2 1608 1624 1309 1368 1013
1 1131 906 1000 1110 1004
0.5 738 446 440 484 431
0.25 343 262 177 244 204
0.125 180 136 122 112 108
0.063 95 71 61 62 58
0.031 40 29 47 25 35](https://image.slidesharecdn.com/comparisonofcommerciallyavailablestrtypingkitsnxpowerlite-13324093356203-phpapp01-120322044250-phpapp01/85/Comparison-Of-Commercially-Available-Str-Typing-Kits-Nx-Power-Lite-9-320.jpg)