Creating a Kinship Matrix Using MSA
- 1. Creating a Kinship Matrix using Microsatellite Analyzer (MSA) Zhifen Zhang The Ohio State University
- 2. Why create a kinship matrix? âĒ Visualize family relatedness âĒ Use in association mapping The Objective of this Tutorial: To introduce one approach to generate a kinship matrix for a population
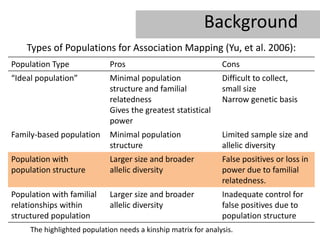
- 3. Background Types of Populations for Association Mapping (Yu, et al. 2006): Population Type Pros Cons âIdeal populationâ Minimal population Difficult to collect, structure and familial small size relatedness Narrow genetic basis Gives the greatest statistical power Family-based population Minimal population Limited sample size and structure allelic diversity Population with Larger size and broader False positives or loss in population structure allelic diversity power due to familial relatedness. Population with familial Larger size and broader Inadequate control for relationships within allelic diversity false positives due to structured population population structure The highlighted population needs a kinship matrix for analysis.
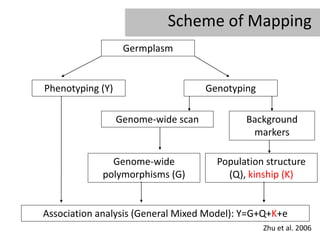
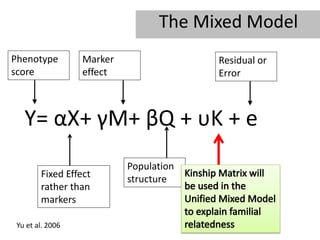
- 4. Scheme of Mapping Germplasm Phenotyping (Y) Genotyping Genome-wide scan Background markers Genome-wide Population structure polymorphisms (G) (Q), kinship (K) Association analysis (General Mixed Model): Y=G+Q+K+e Zhu et al. 2006
- 5. Coefficient of Kinship Coefficient of kinship is used to measure relatedness DEFINITION: Coefficient of kinship is the probability that the alleles of a particular locus chosen randomly from two individuals are identical by descent (Lange, 2002) * Identical by descent: the identical alleles from two individuals arise from the same allele in an earlier generation
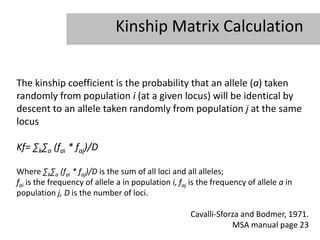
- 6. Kinship Matrix Calculation The kinship coefficient is the probability that an allele (a) taken randomly from population i (at a given locus) will be identical by descent to an allele taken randomly from population j at the same locus Kf= âkâa (fai * faj)/D Where âkâa (fai * faj)/D is the sum of all loci and all alleles; fai is the frequency of allele a in population i, faj is the frequency of allele a in population j, D is the number of loci. Cavalli-Sforza and Bodmer, 1971. MSA manual page 23
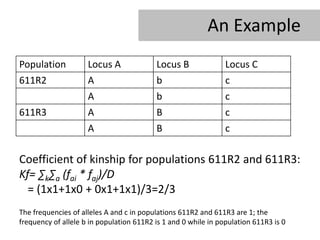
- 7. An Example Population Locus A Locus B Locus C 611R2 A b c A b c 611R3 A B c A B c Coefficient of kinship for populations 611R2 and 611R3: Kf= âkâa (fai * faj)/D = (1x1+1x0 + 0x1+1x1)/3=2/3 The frequencies of alleles A and c in populations 611R2 and 611R3 are 1; the frequency of allele b in population 611R2 is 1 and 0 while in population 611R3 is 0
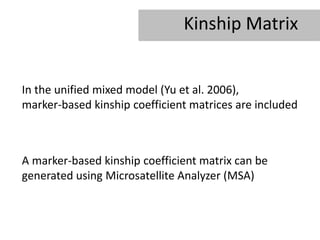
- 8. Kinship Matrix In the unified mixed model (Yu et al. 2006), marker-based kinship coefficient matrices are included A marker-based kinship coefficient matrix can be generated using Microsatellite Analyzer (MSA)
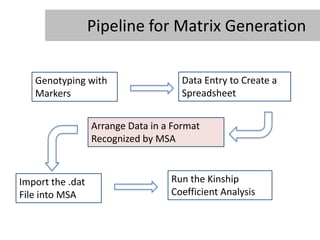
- 9. Pipeline for Matrix Generation Genotyping with Data Entry to Create a Markers Spreadsheet Arrange Data in a Format Recognized by MSA Import the .dat Run the Kinship File into MSA Coefficient Analysis
- 10. A Case Study Bacterial spot data provided by Dr. David Francis, The Ohio State University
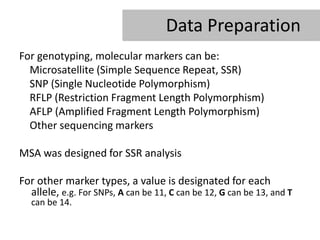
- 11. Data Preparation For genotyping, molecular markers can be: Microsatellite (Simple Sequence Repeat, SSR) SNP (Single Nucleotide Polymorphism) RFLP (Restriction Fragment Length Polymorphism) AFLP (Amplified Fragment Length Polymorphism) Other sequencing markers MSA was designed for SSR analysis For other marker types, a value is designated for each allele, e.g. For SNPs, A can be 11, C can be 12, G can be 13, and T can be 14.
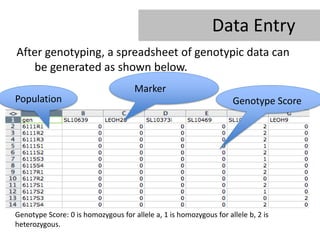
- 12. Data Entry After genotyping, a spreadsheet of genotypic data can be generated as shown below. Marker Population Genotype Score Genotype Score: 0 is homozygous for allele a, 1 is homozygous for allele b, 2 is heterozygous.
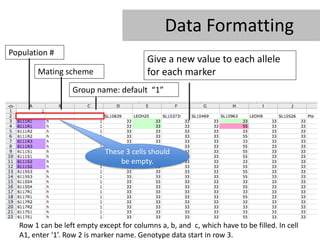
- 13. Data Formatting Population # Give a new value to each allele Mating scheme for each marker Group name: default â1â These 3 cells should be empty. Row 1 can be left empty except for columns a, b, and c, which have to be filled. In cell A1, enter â1â. Row 2 is marker name. Genotype data start in row 3.
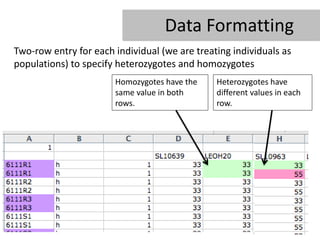
- 14. Data Formatting Two-row entry for each individual (we are treating individuals as populations) to specify heterozygotes and homozygotes Homozygotes have the Heterozygotes have same value in both different values in each rows. row.
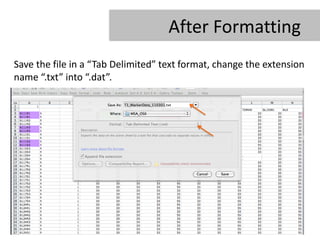
- 15. After Formatting Save the file in a âTab Delimitedâ text format, change the extension name â.txtâ into â.datâ.
- 16. MSA (Microsatellite Analyzer) is available for free, provided by Daniel Dieringer.
- 17. MSA is compatible with different operation systems Choose the operating system you use and install it as other software
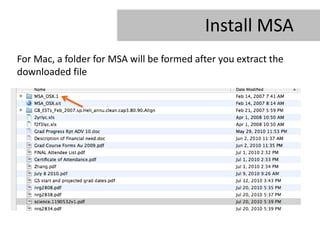
- 18. Install MSA For Mac, a folder for MSA will be formed after you extract the downloaded file
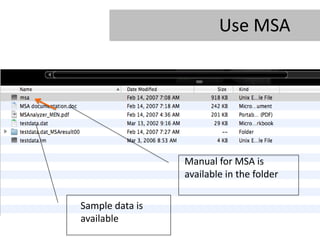
- 19. Use MSA Manual for MSA is available in the folder Sample data is available
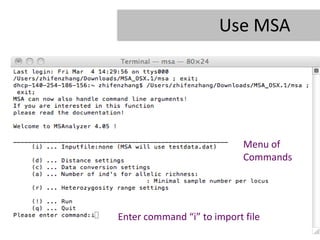
- 20. Use MSA Menu of Commands Enter command âiâ to import file
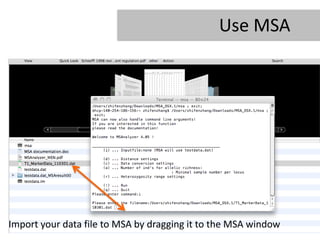
- 21. Use MSA Import your data file to MSA by dragging it to the MSA window
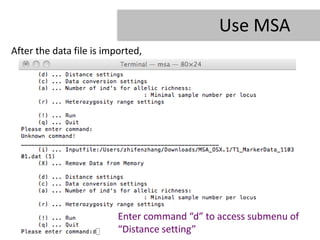
- 22. Use MSA After the data file is imported, Enter command âdâ to access submenu of âDistance settingâ
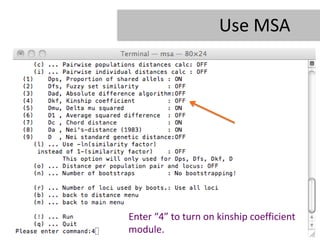
- 23. Use MSA Enter â4â to turn on kinship coefficient module.
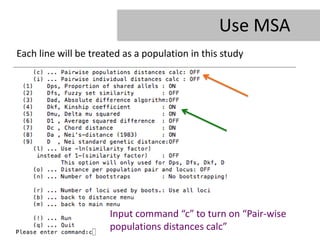
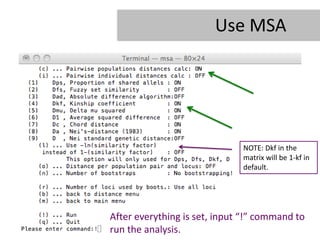
- 24. Use MSA Each line will be treated as a population in this study Input command âcâ to turn on âPair-wise populations distances calcâ
- 25. Use MSA NOTE: Dkf in the matrix will be 1-kf in default. After everything is set, input â!â command to run the analysis.
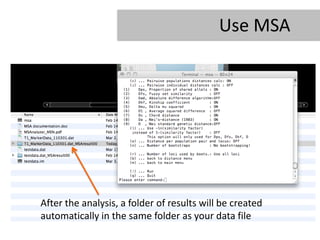
- 26. Use MSA After the analysis, a folder of results will be created automatically in the same folder as your data file
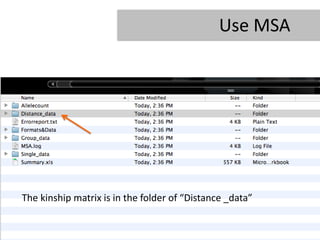
- 27. Use MSA The kinship matrix is in the folder of âDistance _dataâ
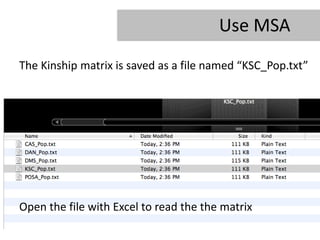
- 28. Use MSA The Kinship matrix is saved as a file named âKSC_Pop.txtâ Open the file with Excel to read the the matrix
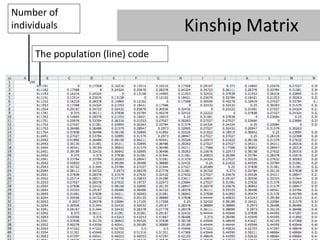
- 29. Number of individuals Kinship Matrix The population (line) code
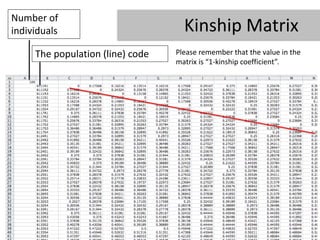
- 30. Number of individuals Kinship Matrix The population (line) code Please remember that the value in the matrix is â1-kinship coefficientâ.
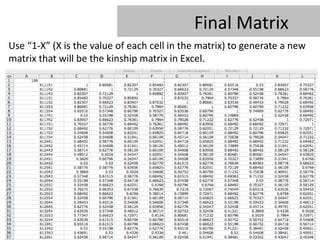
- 31. Final Matrix Use â1-Xâ (X is the value of each cell in the matrix) to generate a new matrix that will be the kinship matrix in Excel.
- 32. The Mixed Model Phenotype Marker Residual or score effect Error Y= ÎąX+ ÎģM+ ÎēQ + Ï K + e Population Fixed Effect structure rather than markers Yu et al. 2006
- 33. References Cited & External Link References Cited âĒ Cavalli-Sforza, L. L., and W. F. Bodmer. 1971. The genetics of human populations, W.H. Freeman and Company, NY. âĒ Lange, K. 2002. Mathematical and statistical methods for genetic analysis, 2nd edition. Springer-Verlag, NY. âĒ Yu, J. G. Pressoir, W. H. Briggs, I. V. Bi, M. Yamasaki, J. F. Doebley, M. D. McMullen, B. S. Gaut, D. M. Nielsen, J. B. Holland, S. Kresovich, and E. S. Buckler. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38: 203-208. (Available online at: http://dx.doi.org/10.1038/ng1702) (verified 8 July 2011). âĒ Zhu, C. M. Gore, E. S. Buckler, and J. Yu. 2008. Status and prospects of association mapping in plants. The Plant Genome 1: 5-20. (Available online at: http://dx.doi.org/10.3835/plantgenome2008.02.0089) (verified 8 July 2011). External Link âĒ Dieringer, D. Microsatellite analyzer (MSA) [Online]. Available at: http://i122server.vu-wien.ac.at/MSA/MSA_download.html (verified 8 July 2011).




![References Cited & External Link
References Cited
âĒ Cavalli-Sforza, L. L., and W. F. Bodmer. 1971. The genetics of human
populations, W.H. Freeman and Company, NY.
âĒ Lange, K. 2002. Mathematical and statistical methods for genetic analysis, 2nd
edition. Springer-Verlag, NY.
âĒ Yu, J. G. Pressoir, W. H. Briggs, I. V. Bi, M. Yamasaki, J. F. Doebley, M. D.
McMullen, B. S. Gaut, D. M. Nielsen, J. B. Holland, S. Kresovich, and E. S.
Buckler. 2006. A unified mixed-model method for association mapping that
accounts for multiple levels of relatedness. Nature Genetics 38: 203-208.
(Available online at: http://dx.doi.org/10.1038/ng1702) (verified 8 July 2011).
âĒ Zhu, C. M. Gore, E. S. Buckler, and J. Yu. 2008. Status and prospects of
association mapping in plants. The Plant Genome 1: 5-20. (Available online at:
http://dx.doi.org/10.3835/plantgenome2008.02.0089) (verified 8 July 2011).
External Link
âĒ Dieringer, D. Microsatellite analyzer (MSA) [Online]. Available at:
http://i122server.vu-wien.ac.at/MSA/MSA_download.html (verified 8 July
2011).](https://image.slidesharecdn.com/kinshipmatrixfinal-120402144019-phpapp02/85/Creating-a-Kinship-Matrix-Using-MSA-33-320.jpg)