Diagnostic hypothesis refinement in reproducible workflows for advanced medical data analysis
- 1. Diagnostic hypothesis refinement in reproducible workflows for advanced medical data analysis Cezary Mazurek, Raul Palma, Juliusz Pukacki Pozna©Į Supercomputing and Networking Center Scientific workshop. Big Data: processing and exploration, 22.04.2016, Pozna©Į,
- 2. Workflows ?? The automation of a business process, in whole or part, during which documents, information or tasks are passed from one participant to another for action, according to a set of procedural rules. (From The Workflow Management Coalition Specification) ?? Workflows serve a dual function *): ©C? first as detailed documentation of the method (i. e. the input sources and processing steps taken for the derivation of a certain data item) ©C? second as re-usable, executable artifacts for data-intensive analysis. ?? Workflows stitch together a variety of data manipulation activities such as data movement, data transformation or data visualization to serve the goals of the scientific study*). *) D.Garijo,P.Alper,K.Belhajjame,O.Corcho,Y.Gil,C.Goble,Common motifs in scientific workflows: an empirical analysis, Future Gener. Comput. Syst.(2014) http://dx.doi.org/10.1016/j.future.2013.09.018.
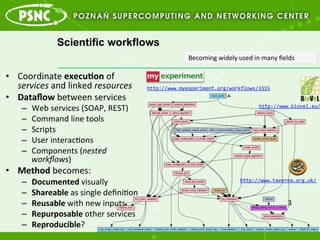
- 3. Scientific workflows ?? Coordinate ?execu%on ?of ? services ?and ?linked ?resources ? ?? Data?ow ?between ?services ? ©C? Web ?services ?(SOAP, ?REST) ? ©C? Command ?line ?tools ? ©C? Scripts ? ©C? User ?interacAons ? ©C? Components ?(nested ? work?ows) ? ?? Method ?becomes: ? ©C? Documented ?visually ? ©C? Shareable ?as ?single ?de?niAon ? ©C? Reusable ?with ?new ?inputs ? ©C? Repurposable ?other ?services ? ©C? Reproducible? ? http://www.myexperiment.org/workflows/3355 ? http://www.taverna.org.uk/ ? http://www.biovel.eu/ 3 ? Becoming ?widely ?used ?in ?many ??elds ?
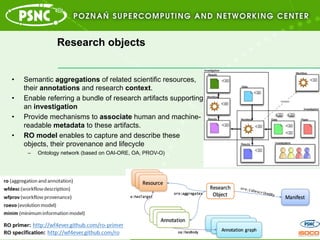
- 4. Research objects ?? Semantic aggregations of related scientific resources, their annotations and research context. ?? Enable referring a bundle of research artifacts supporting an investigation ?? Provide mechanisms to associate human and machine- readable metadata to these artifacts. ?? RO model enables to capture and describe these objects, their provenance and lifecycle ©C? Ontology network (based on OAI-ORE, OA, PROV-O)
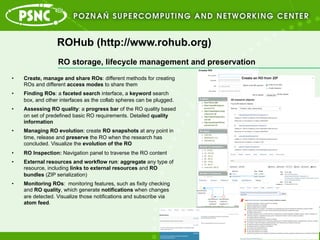
- 5. ROHub (http://www.rohub.org) ?? Enables the sharing of scientific findings ?? Support scientists throughout the research lifecycle to create and maintain high-quality ROs that can be interpreted and reproduced in the future. ?? Combination of digital libraries, long term-preservation and semantic technologies. RO storage, lifecycle management and preservation
- 6. ROHub (http://www.rohub.org) ?? Create, manage and share ROs: different methods for creating ROs and different access modes to share them ?? Finding ROs: a faceted search interface, a keyword search box, and other interfaces as the collab spheres can be plugged. ?? Assessing RO quality: a progress bar of the RO quality based on set of predefined basic RO requirements. Detailed quality information ?? Managing RO evolution: create RO snapshots at any point in time, release and preserve the RO when the research has concluded. Visualize the evolution of the RO ?? RO Inspection: Navigation panel to traverse the RO content ?? External resources and workflow run: aggregate any type of resource, including links to external resources and RO bundles (ZIP serialization) ?? Monitoring ROs: monitoring features, such as fixity checking and RO quality, which generate notifications when changes are detected. Visualize those notifications and subscribe via atom feed. RO storage, lifecycle management and preservation
- 7. Reproducibility Reproducibility for computational experiments is challenging. It is hard both for authors to derive a compendium that encapsulates all the components (e.g., data, code, parameter settings, environment) needed to reproduce a result, and for reviewers to verify the results. There are also other barriers, from practical issues ©C including the use of proprietary data, software and specialized hardware, to social ©C for example, the lack of incentives for authors to spend the extra time making their experiments reproducible. Challenge
- 9. T.Marschal: ?In ?Vivo, ?in ?vitro, ?in ?Silico!. ?ANSYS ?Advantage, ?vol. ?IX, ?Issue ?1, ?2015 ?
- 10. Problem/Challenge ?? Historically, the scientific method is well known and was introduced by Louis Pasteur in XIX century. ?? This method is in fact a cycle of following steps: ©C? Observations->Questions->Hypotheses-> Predictions>Experiment (incl. refinement) -> Discussion. ?? These steps allowed for many years to report scientific experiments conducted In-Vivo and In-Vitro. ?? However we think that even if steps are still the same while performing in-Silico experiments, the way of reporting them need to be changed, especially in fields where part of experiment is creation of software tools
- 11. What it means? ?? Smart data processing and experients butĪŁ. ?? What data means for doctors? ?? They need treatment instructions and its expected results ?? We need new environment for in-silico disease hypothesis refinement and building decision support systems This is a challenge for researchers in interdisciplinary teams
- 12. Prof ?Mark ?Caul?eld ?FMedSci, ?Genomics ?England ?Clinical ?InterpretaAon ?Partnership ?
- 13. Are the answers obvious? Are the questions obvious?
- 14. Towards precision (personal) medicine ?? Questions-driven (smart) data experiments ?? If failed ©C lead to other questions and experiments ?? SoĪŁdo not start from transfering existing knowledge and statistical approach to data space ?? We need to start thinking from like being lived in data space and create experiments to quickly verify hypothesis (diagnostic hypothesis refinement) ?? Precision medicine makes it even more challenging!!! ©C? Data experiments are being defined for individual patient and route to personal treatment
- 15. Disruptive Innovation in Interdisciplinary Teams Decision ?support ?systems ?for ?disease ?diagnosis ? Diagnos%c ?hypothesis ? ?re?nement ? Smart ?processing ? Data ?
- 17. Hypothesis refinement ?? In-Silico experiments, especially in their refinement cycle, lead to creation of new software tools, algorithms and even computer science challenges. To make this experiment valuable such a process needs to be controlled and recorded while achieving milestone stages; ?? Scientific experiments are performed in cycles, when each cycle is a refinement of the hypothesis. Continuing research starting from any cycle and branching this process further on, require that each cycle is checkpointed and stored as a scientific procedure step; ?? Medical research reliant on data analysis, focused on early disease diagnosis or stopping the disease progress, very often results in providing software tools helping in data analysis and created during the experimentation cycles. ?? To treat the process of knowledge discovery based on data analysis and development of processing tools, as a research method, we need to provide the way of formal description of stages of such a process, be paired with hypothesis refinement stages. Practical cases
- 18. Domain examples ?? Bioinformatics ©C? *omics research ?? Earth Science (EVEREST) ©C? European Virtual Environment for Research - Earth Science Themes: a solution ?? Cardiac rehabilitation and early risk identification of cardiovasular diseases ©C? Personal prevention plan ?? Glaucoma diagnosis and early prevention
- 19. Glaucoma research experiment Glaucoma - group of progressive optic nerve neuropaties releted with: a) accelerated apoptosis of Retinal Ganglion Cells due to neurotrophic deprivation [Band L.R., 2009; Balaratnasingam C., 2008; Fechtner R.D., Weinreb R.N., 1994; Garcia- Valenzuela E., 1995; Quigley H.A., 1976, 1995, 2000; Yablonski M., Asamoto A., 1993] b)?lamina cribrosa sclerae pathognomonic phenotype changes [Ernest J.T. and Potts A.M., 1968; Quigley H.A., 1983; Roberts M.D., 2009].
- 20. StepsANALIZA DANYCH ©C INTERWA?Y CZASOWE 0 ? 50 ? 100 ? 150 ? 200 ? 250 ? 13.00 ? 13.30 ? 14.00 ? 14.30 ? 15.00 ? 15.30 ? 16.00 ? 16.30 ? 17.00 ? 17.30 ? 18.00 ? 18.30 ? 19.00 ? 19.30 ? 20.00 ? 20.30 ? 21.00 ? 21.30 ? 22.00 ? 23.00 ? 0.00 ? 1.00 ? 2.00 ? 3.00 ? 4.00 ? 5.00 ? 6.00 ? 7.00 ? 7.30 ? 8.00 ? 8.30 ? 9.00 ? 9.30 ? 10.00 ? 10.30 ? 11.00 ? 11.30 ? 12.00 ? 12.30 ? 13.00 ? SAP ? DAP ? 0 ? 50 ? 100 ? 150 ? 200 ? HR ?BP ? 1.? GENERAL ANALYSIS - AREA UNDER CURVE (AUC) 24h 2.? TIME-INTERVAL DEPENDENT ANALYSIS (Linear Model ”┴ & ”┬) 4 ?3 ?2 ?1 ? TFĪ▒0Ī▒
- 21. Checkpoint ?? ?280 ?rules ?assigned ?into ?50 ?classi?ers ? ?( ?role ?of ?Experts) ? ?? ?Classi?ers ?VoAng ?( ?round ?table) ?decide ?of ?diagnosis ? ?? ?Rules ?indicated ?by ?algorithm ?in ?diagnosis ? ?pointed ?at ?speci?c ?place ?of ?pathology ? ? ?in ?checked ?system? ? Decision ?Rule ?Models ?in ?Di?erenAaAon ?of ?Healthy ?and ?Glaucomatous ?PaAentsĪ▒ ?R. ?Wasilewicz; ?P. ?Wasilewicz; ?A. ?Radziemski, ?J. ?B?aszczy©Įski, ? C. ?Mazurek; ?R. ?S?owinski, ?Cardiovascular ?Mobile ?Health ?Conference ?2015, ?Tabarz, ?Germany ?
- 22. Hypothesis Experiment ? Stage Processing Data ?Space ? Exp Result Stage Result Dataset Preprocessing
- 23. Interna%onal ?Consor%um ? Open ?Health ?System ? Laboratory, ?USA ? University ?of ?Notre ? Dame, ?USA ? Internet2, ?USA ? Centre ?for ?Development ? of ?Advanced ?CompuAng, ? India ? Chalmers ?Unviersity ?of ? Technology, ?Sweden ? Pozna©Į ?SupercompuAng ? and ?Networking ?Center, ? Poland ? Indian ?InsAtute ?of ? Technology, ?Dehli, ?India ? ? Duke ?University ?Ī¬ ?Applied ? TherapeuAcs ?SecAon, ?USA ? In ?collabora%on ?with: ?
- 24. Interna%onal ?collabora%on ?for ?biomedicine ?
- 25. Applica%ons ?(some ?examples) ? CDAC ? ? Biomolecular ?SimulaAons ?and ? molecular ?docking: ?Research ?on ? cancer ?proteins, ?anAsense ? molecules, ?GPCRs ? ? Next ?GeneraAon ?Sequencing ?Data ? Analysis: ?ApplicaAons ?in ?cancer ? genomics ?(Breast ?Cancer ? transcriptome) ? ? High ?throughput ?comparaAve ? genomics ?studies ?on ?salmonella ? and ?mycobacterium ? ? Chalmers ? ? Chalmers ?Life ?Science ?and ? Engineering: ? ?EuropeĪ»s ?leading ? center ?for ?Metabolic ?Engineering ? and ?Systems ?Biology ?(Jens ?Nielsen ? Lab) ? ? Gothenburg ?University ?(Molecular ? Biology, ?EuropeĪ»s ?leading ?Center ? for ?Systems ?Biology,NGS) ? ? Sahlgrenska ?University ?Hospital ? and ?Academy ?(Centers ?for ?Cancer ? and ?Cardiovascular ?and ?Metabolic ? Diseases) ? ? Biotech ?Industries: ?AstraZeneca ? worldwide ?research ?and ? innovaAon ?hub. ? ? PSNC ? ? Support ?for ?complex ?eScience ? research ?tasks ?in ?the ?area ?of ?post-?®\ genomic ?clinical ?trials ?and ?virtual ? physical ?human ?modeling ?for ? clinical ?purposes: ?ACGT ?and ?P-?®\ Medicine ?projects ? ? RNASeq ?analysis ?(role ?of ?proteins ? and ?retroelements ?in ?induced ? pluripotent ?stem ?cells) ? ? Breast ?cancer ?therapy ?(novel ? biomarkers) ?and ?diagnosAcs ? (applying ?TCGA ?data) ? ? InteracAve ?visualizaAon ?of ? correlaAons ?between ?genomic ? analysis ?observaAons ? Pilot ?work?ow ?integraAon ?with ? UT ?MD ?Anderson ?Cancer ?Center ?
- 26. GEN Exclusive We need new models for collaboration between the health research industry and academia. The only way that will happen is if we can reduce some of the local competition and fragmentation and create super-centers of innovation for: ?? regional consortia for clinical research, ?? experimental therapeutics centers, ?? advanced biomanufacturing centers, ?? centralized repositories for patient data. hpp://leadership.je?erson.edu/blog/ ?
- 28. Publications ?? R.Wasilewicz, P.Wasilewicz, E.Czaplicka, J.KocieckiI, J.Blaszczynski, C.Mazurek and R.Slowinski: 24 hour continuous ocular tonography Triggerfish and biorhythms of the cardiovascular system functional parameters in healthy and glaucoma populations. Acta Ophthalmologica, 91: 0. doi: 10.1111/j.1755-3768.2013.2721.x ?? Palma R., Corcho O., Ho?ubowicz P., P©”rez S., Page K., Mazurek C., Digital libraries for the preservation of research methods and associated artefacts. Proc. 1st International Workshop on the Digital Preservation of Research Methods and Artefacts (DPRMA 2013) at Joint Conference on Digital Libraries (JCDL 2013). pp. 8-15. Indianapolis, Indiana, USA, July 2013 ?? Mazurek, C., Pukacki, J., Kosiedowski, M., Trocha, S., Darbari, H., Saxena, A., Joshi, R., Brenner, P., Gesing, S., Nabrzyski, J., Sullivan, M., Dubhashi, D., Thankaswamy, S., and Srivastava, A. (2014) Federated Clouds for Biomedical Research: Integrating OpenStack for ICTBioMed. Cloud Networking (CloudNet), 2014 IEEE 3rd International Conference on, pp.294-299, 8-10 Oct. 2014, doi: 10.1109/CloudNet.2014.6969011 ?? Palma R., Corcho O., G©«mez-P©”rez J.M., Mazurek, C., Ī░ROHub A Digital Library of Research Objects Supporting Scientists Towards Reproducible ScienceĪ▒. In Semantic Publishing Challenge of Proc. Extended Semantic Web Conference (ESWC), Crete, Greece, May 25-29, 2014. ?? M.Krysinski, M.Krystek, C.Mazurek, J.Pukacki, P.Spychala, M.Stroinski, J.Weglarz. Semantic Data Sharing and Presentation in Integrated Knowledge System. [In:] R. Bembenik, ?. Skonieczny, H. Rybi©Įski, M. Kryszkiewicz, & M. Niezg©«dka (Eds.), Intelligent Tools for Building a Scientific Information Platform: Advanced Architectures and Solutions, pp. 67©C83. Springer International Publishing 2013 ?? J.Andersen, P.Shah, K.Korski, M.Ibbs, V.Filas, M.Kosiedowski, J.Pukacki, C.Mazurek, Y.Wu, E.Chang, C.Toniatti, G.Draetta, M.Wiznerowicz: Applying TCGA data for breast cancer diagnostics and pathway analysis, Cancer Research 10/2014; 74(19 Supplement):4272-4272 ?? J.Pukacki, H.?wierczy©Įski, C.Mazurek, M.Kosiedowski "RNA-Seq data analysis pipeline in Poznan Supercomputing and Networking Center", 1st Congress of the Polish Biochemistry, Cell Biology, Biophysics and Bioinformatics, September 2014, Warsaw, Poland ?? M. Kosiedowski, C. Mazurek, K. S?owi©Įski, M. Stroi©Įski, K. Szyma©Įski, J. W?glarz: ?Telemedical systems for the support of regional Healthcare In the area of traumaĪ▒ , Global Telemedicine and Health Updates: Knowledge Resources, vol. 3 str. 592 ©C 596, 2010
- 29. Pozna©Į Supercomputing and Networking Center ul. Noskowskiego 12/14, 61-704 Pozna©Į, POLAND, Office: phone center: (+48 61) 858-20-00, fax: (+48 61) 852-59-54, e-mail: office@man.poznan.pl, http://www.psnc.pl affiliated to the Institute of Bioorganic Chemistry of the Polish Academy of Sciences,















![Glaucoma research experiment
Glaucoma - group of progressive optic nerve neuropaties releted with:
a) accelerated apoptosis of Retinal Ganglion Cells due to neurotrophic deprivation
[Band L.R., 2009; Balaratnasingam C., 2008; Fechtner R.D., Weinreb R.N., 1994; Garcia-
Valenzuela E., 1995; Quigley H.A., 1976, 1995, 2000; Yablonski M., Asamoto A., 1993]
b)?lamina cribrosa sclerae pathognomonic phenotype changes
[Ernest J.T. and Potts A.M., 1968; Quigley H.A., 1983; Roberts M.D., 2009].](https://image.slidesharecdn.com/mazurek-2016-04-22-bigdata-www-160515154213/85/Diagnostic-hypothesis-refinement-in-reproducible-workflows-for-advanced-medical-data-analysis-19-320.jpg)








![Publications
?? R.Wasilewicz, P.Wasilewicz, E.Czaplicka, J.KocieckiI, J.Blaszczynski, C.Mazurek and R.Slowinski: 24 hour continuous ocular
tonography Triggerfish and biorhythms of the cardiovascular system functional parameters in healthy and glaucoma populations.
Acta Ophthalmologica, 91: 0. doi: 10.1111/j.1755-3768.2013.2721.x
?? Palma R., Corcho O., Ho?ubowicz P., P©”rez S., Page K., Mazurek C., Digital libraries for the preservation of research methods and
associated artefacts. Proc. 1st International Workshop on the Digital Preservation of Research Methods and Artefacts (DPRMA
2013) at Joint Conference on Digital Libraries (JCDL 2013). pp. 8-15. Indianapolis, Indiana, USA, July 2013
?? Mazurek, C., Pukacki, J., Kosiedowski, M., Trocha, S., Darbari, H., Saxena, A., Joshi, R., Brenner, P., Gesing, S., Nabrzyski, J.,
Sullivan, M., Dubhashi, D., Thankaswamy, S., and Srivastava, A. (2014) Federated Clouds for Biomedical Research: Integrating
OpenStack for ICTBioMed. Cloud Networking (CloudNet), 2014 IEEE 3rd International Conference on, pp.294-299, 8-10 Oct.
2014, doi: 10.1109/CloudNet.2014.6969011
?? Palma R., Corcho O., G©«mez-P©”rez J.M., Mazurek, C., Ī░ROHub A Digital Library of Research Objects Supporting Scientists
Towards Reproducible ScienceĪ▒. In Semantic Publishing Challenge of Proc. Extended Semantic Web Conference (ESWC), Crete,
Greece, May 25-29, 2014.
?? M.Krysinski, M.Krystek, C.Mazurek, J.Pukacki, P.Spychala, M.Stroinski, J.Weglarz. Semantic Data Sharing and Presentation in
Integrated Knowledge System. [In:] R. Bembenik, ?. Skonieczny, H. Rybi©Įski, M. Kryszkiewicz, & M. Niezg©«dka (Eds.), Intelligent
Tools for Building a Scientific Information Platform: Advanced Architectures and Solutions, pp. 67©C83. Springer International
Publishing 2013
?? J.Andersen, P.Shah, K.Korski, M.Ibbs, V.Filas, M.Kosiedowski, J.Pukacki, C.Mazurek, Y.Wu, E.Chang, C.Toniatti, G.Draetta,
M.Wiznerowicz: Applying TCGA data for breast cancer diagnostics and pathway analysis, Cancer Research 10/2014; 74(19
Supplement):4272-4272
?? J.Pukacki, H.?wierczy©Įski, C.Mazurek, M.Kosiedowski "RNA-Seq data analysis pipeline in Poznan Supercomputing and
Networking Center", 1st Congress of the Polish Biochemistry, Cell Biology, Biophysics and Bioinformatics, September 2014,
Warsaw, Poland
?? M. Kosiedowski, C. Mazurek, K. S?owi©Įski, M. Stroi©Įski, K. Szyma©Įski, J. W?glarz: ?Telemedical systems for the support of
regional Healthcare In the area of traumaĪ▒ , Global Telemedicine and Health Updates: Knowledge Resources, vol. 3 str. 592 ©C
596, 2010](https://image.slidesharecdn.com/mazurek-2016-04-22-bigdata-www-160515154213/85/Diagnostic-hypothesis-refinement-in-reproducible-workflows-for-advanced-medical-data-analysis-28-320.jpg)
