dmontaner at cipf_2014
- 1. The road of excess . . . leads to the palace of wisdom (William Blake - 1790) David Montaner 2014-06-19 David Montaner Computational Genomics @ CIPF 1/15
- 2. About Statistician working in genomic research Computational Genomics Department (Bioinformatics - Ximo Dopazo) Massive (high-throughput) data analysis - Genomics methods software and tools practical application Teaching . . . bioStat-coaching short courses in bioinformatics University of Valencia student tutoring e-learning materials CEO at Genometra: bioinformatics consultancy David Montaner Computational Genomics @ CIPF 2/15
- 3. Publications 2013 Di?erential gene-expression analysis de?nes a molecular pattern related to olive pollen allergy. Aguerri M, Calzada D, Montaner D, Mata M, Florido F, Quiralte J, Dopazo J, Lahoz C, Cardaba B. J Biol Regul Homeost Agents. 2013 Apr-Jun;27(2):337-50. De?ning the genomic signature of totipotency and pluripotency during early human development. Galan A, Diaz-Gimeno P, Poo ME, Valbuena D, Sanchez E, Ruiz V, Dopazo J, Montaner D, Conesa A, Simon C. PLoS One. 2013 Apr 17;8(4):e62135. doi: 10.1371/journal.pone.0062135. Print 2013. Thesis Methodological advances in the functional pro?ling of genomic studies. David Montaner Computational Genomics @ CIPF 3/15
- 4. Publications 2014 A novel locus for a hereditary recurrent neuropathy on chromosome 21q21. E Calpena, D MartĻŠnez-Rubio, J Arpa, JJ GarcĻŠa-Pe?as, D Montaner, J Dopazo, F Palau, C EspinĻŪs. Neuromuscular Disorders, 2014. Pathway network inference from gene expression data. Ignacio Ponzoni, MarĻŠa JosĻĶ Nueda, Sonia Tarazona, Stefan G?tz, David Montaner, Julieta Sol Dussaut, JoaquĻŠn Dopazo, Ana Conesa. BMC Systems Biology, 2014. 8 (2), 1-17. Quantitative modeling of clinical, cellular, and extracellular matrix variables suggest prognostic indicators in cancer: a model in neuroblastoma. Tadeo I, Piqueras M, Montaner D, VillamĻŪn E, Berbegall AP, Ca?ete A, Navarro S, Noguera R. Pediatr Res. 2014 Feb;75(2):302-14. David Montaner Computational Genomics @ CIPF 4/15
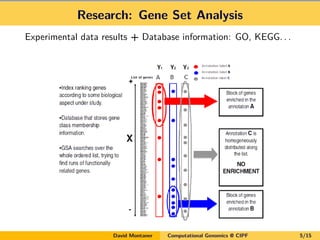
- 5. Research: Gene Set Analysis Experimental data results + Database information: GO, KEGG. . . David Montaner Computational Genomics @ CIPF 5/15
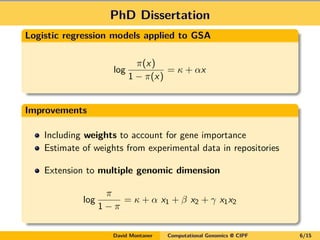
- 6. PhD Dissertation Logistic regression models applied to GSA log ĶÐ(x) 1 ? ĶÐ(x) = ĶĘ + ĶÁx Improvements Including weights to account for gene importance Estimate of weights from experimental data in repositories Extension to multiple genomic dimension log ĶÐ 1 ? ĶÐ = ĶĘ + ĶÁ x1 + ĶÂ x2 + ĶÃ x1x2 David Montaner Computational Genomics @ CIPF 6/15
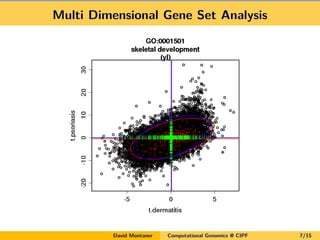
- 7. Multi Dimensional Gene Set Analysis David Montaner Computational Genomics @ CIPF 7/15
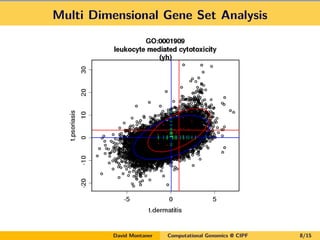
- 8. Multi Dimensional Gene Set Analysis David Montaner Computational Genomics @ CIPF 8/15
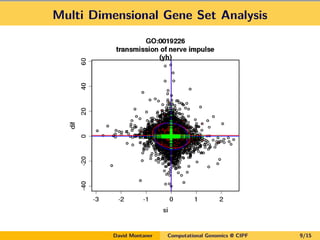
- 9. Multi Dimensional Gene Set Analysis David Montaner Computational Genomics @ CIPF 9/15
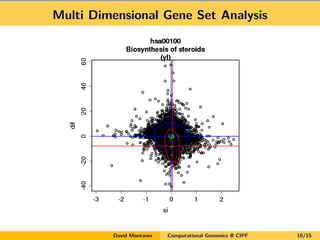
- 10. Multi Dimensional Gene Set Analysis David Montaner Computational Genomics @ CIPF 10/15
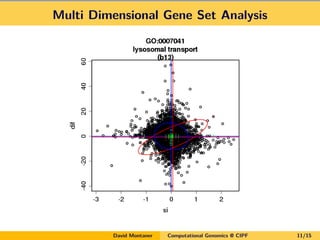
- 11. Multi Dimensional Gene Set Analysis David Montaner Computational Genomics @ CIPF 11/15
- 12. Ongoing Work Improving software implementation: R package, Babelomics tool Adjust the methods to NGS data Extend to genomic features other than genes miRNAs SNPs Functional Meta-analysis: combine GSA results from several datasets New research interests: Topological pathway analysis Metagenomics: genomic analysis of microbial communities David Montaner Computational Genomics @ CIPF 12/15
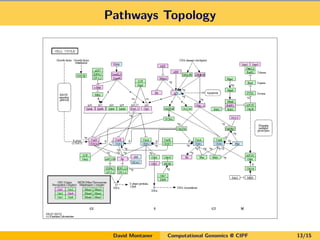
- 13. Pathways Topology David Montaner Computational Genomics @ CIPF 13/15
- 14. Ongoing work Improving software implementation: R package, Babelomics Tool Adjust the methods to NGS data Extend to genomic features other than genes miRNAs SNPs Functional Meta-analysis: combine GSA results from several datasets New research interests: Topological pathway analysis Metagenomics: genomic analysis of microbial communities David Montaner Computational Genomics @ CIPF 14/15
- 15. Thank You Further reading . . . www.dmontaner.com bioinfo.cipf.es/dmontaner www.linkedin.com/in/dmontaner twitter.com/david_montaner David Montaner Computational Genomics @ CIPF 15/15