Exchange program 071128
0 likes78 views
Exchange Program, UNIVERSITY OF THE PHILIPPINES LOS BA?OS
1 of 12
Download to read offline











Ad
Recommended
ChatGPT ż╬¼Fū┤└ĒĮŌż╚ 2023─Ļ7į┬░µ LLMŪķł¾źóź├źūźŪ®`ź╚
ChatGPT ż╬¼Fū┤└ĒĮŌż╚ 2023─Ļ7į┬░µ LLMŪķł¾źóź├źūźŪ®`ź╚Satoshi Kume
?
1. ChatGPTż╬¼Fū┤└ĒĮŌ
ĪĪChatGPT ż╚żŽŻ┐
ĪĪŪ░╗žż╬š±żĻĘĄżĻ
ĪĪChatGTPĮń┌±ż╬1ź÷į┬ķgż╬źóź├źūźŪ®`ź╚ (2023─Ļ7į┬░µ)
2. Rźčź├ź▒®`źĖķ_░kż╦LLMż“ż╔ż”╗Ņė├ż╣żļż½Ż┐ż╬š¹└Ē
3. GPTż¼╩╣ż©żļRźčź├ź▒®`źĖż╬š{¢╦ł¾Ėµ
230517_chatGPT_v01.pdf
230517_chatGPT_v01.pdfSatoshi Kume
?
##ChatGPTż╬¼Fū┤└ĒĮŌż╚Rķv╩²&źčź├ź▒®`źĖū„│╔żžż╬╗Ņė├
1. ChatGPTż╬¼Fū┤└ĒĮŌ
OpenAI╔ńż╦ż─żżżŲ
ChatGPTż╚żŽŻ┐
GPT-3.5ż╚GPT-4
źšźĒź¾źūź╚ż╚żŽŻ┐
ChatGPTż╬Ą├ęŌż╩ż│ż╚?┐Ó╩ųż╩ż│ż╚Ż©╩┬└²ż╚TipsżŌŻ®
2. Rķv╩²&źčź├ź▒®`źĖū„│╔żžż╬╗Ņė├
GPT API keyż╬╚ĪĄ├
gptstudioźčź├ź▒®`źĖż“╩╣ż├żŲĪóRStudio╔ŽżŪChatGPTż“╩╣ė├ż╣żļ╩┬└²ż╬ĮBĮķ211104 Bioc Asia workshop
211104 Bioc Asia workshopSatoshi Kume
?
211104 @ BioC Asia 2021 Workshop
Introduction to Bioimage Analysis in R
This workshop covers basic methods of the image processing and image analysis in R using the Bioconductor package Ī░EBImageĪ▒ and the Orchestra platform. In addition, the image dataset is obtained from ExperimentHub using the Ī░BioImageDbsĪ▒ package. Using this dataset, we perform a supervised image segmentation using the U-NET model, one of deep learning models, provided by the rMiW package.
ż│ż╬ź’®`ź»źĘźńź├źūżŪżŽĪóBioconductorźčź├ź▒®`źĖżŪżóżļEBImageż“╩╣ż├żŲĪóRżŪż╬╗ŁŽ±äI└Ē?╗ŁŽ±ĮŌ╬÷ż╬╗∙▒ŠĄ─ż╩ĘĮĘ©ż“ÆQż”ĪŻ┤╬ż╦ĪóBioImageDbsźčź├ź▒®`źĖż“ė├żżżŲĪóExperimentHubż½żķż╬╗ŁŽ±źŪ®`ź┐ż╬╚ĪĄ├ż“ąąż”ĪŻżĄżķż╦ĪórMiWźčź├ź▒®`źĖż¼╠ß╣®ż╣żļĪóDeep learningźŌźŪźļż╬Ż▒ż─żŪżóżļU-NETźŌźŪźļż“ė├żżżŲĪóĮ╠ĤėążĻ╗ŁŽ±ź╗ź░źßź¾źŲ®`źĘźńź¾Ż©ŅIė“ĘųĖŅŻ®ż“覿ėĪŻż│ż╬ź’®`ź»źĘźńź├źūżŽĪóOrchestraŁhŠ│ż╦żŲīg╩®ż╣żļĪŻCRANźčź├ź▒®`źĖż╬ū„│╔?═ČĖÕż╚ź│®`ź╔źņźėźÕ®`īØÅĻ ? GoogleImage2Array źčź├ź▒®`źĖżŪż╬╩┬└²ĮBĮķ ? @ BioPackathon
CRANźčź├ź▒®`źĖż╬ū„│╔?═ČĖÕż╚ź│®`ź╔źņźėźÕ®`īØÅĻ ? GoogleImage2Array źčź├ź▒®`źĖżŪż╬╩┬└²ĮBĮķ ? @ BioPackathonSatoshi Kume
?
CRANźčź├ź▒®`źĖż╬ū„│╔?═ČĖÕż╚ź│®`ź╔źņźėźÕ®`īØÅĻ
? GoogleImage2Array źčź├ź▒®`źĖżŪż╬╩┬└²ĮBĮķ ?
@BioPackathonD3 slides (Satoshi Kume)
D3 slides (Satoshi Kume)Satoshi Kume
?
źĻź▌ź½źĻź¾ą═źūźĒź╣ź┐ź░źķź¾źĖź¾D║Ž│╔Į═╦žż╬ų¼╚▄ąįĄ═Ęųūėż╦īØż╣żļŽĄĮyĄ─ŽÓ╗źū„ė├ĮŌ╬÷
Systematic Interaction Analysis of Human Lipocalin-type Prostaglandin D Synthase with Small Lipophilic LigandsAIč¦╗ß ║Ž═¼čąŠ┐╗ß2020 ░k▒Ēź╣źķźżź╔ (201120)
AIč¦╗ß ║Ž═¼čąŠ┐╗ß2020 ░k▒Ēź╣źķźżź╔ (201120)Satoshi Kume
?
Ė▀Ęųūė▓─┴Žź¬ź¾ź╚źĒźĖ®`ż╬śŗ║Bż╦Ž“ż▒ż┐ Wikidataż½żķż╬ź╔źßźżź¾Ė┼─Ņ│ķ│÷
Extraction of Domain Concepts from Wikidata for Construction of Polymer Material Ontology210609 Biopackthon: BioImageDbs for ExperimentalHub (ą▐š²░µ)
210609 Biopackthon: BioImageDbs for ExperimentalHub (ą▐š²░µ)Satoshi Kume
?
BioImageDbs (for ExperimentHub)ż╬ķ_░kż╚żĮż│ż½żķ覿¾ż└źčź├ź▒®`źĖū„│╔ż╬Tipsż╦ż─żżżŲOpenlink Virtuoso v01
Openlink Virtuoso v01Satoshi Kume
?
ĪĖOpenlink VirtuosoĪ╣ż╬źżź¾ź╣ź╚®`źļż╚įOČ©ż╬╚š▒ŠšZ┘Y┴Ž
?Wikitadaź└ź¾źūż╬╚ĪĄ├
?Mac ░µ Virtuoso ż╬źżź¾ź╣ź╚®`źļż╚įOČ©
?Win░µ Virtuoso ż╬źżź¾ź╣ź╚®`źļż╚įOČ©
201209 Biopackathon 12th
201209 Biopackathon 12thSatoshi Kume
?
╗ŁŽ±äI└Ē?╗ŁŽ±ĮŌ╬÷ż╦ķvż╣żļRźčź├ź▒®`źĖż“š{¢╦żĘż┐ż╬żŪĪóęŖż─ż½ż├ż┐źčź├ź▒®`źĖż“Ż©Ä┌ż»Ū│ż»Ż®ĮBĮķż╣żļĪŻ
š{¢╦īØŽ¾: CRANĪóBioconductorĪóNeuroconductor (= GitHub/CRAN)ż╚ż½
Ś╩╦„źŁ®`ź’®`ź╔: imag(e), microscop(y) ctc
ANTsRNetż“╩╣ż├żŲĪó2D U-NET/CPU░µż“żõż├żŲż▀ż┐ĪŻ201126 Wikidata ź»źķź╣ļAīė SPARQLŚ╩╦„
201126 Wikidata ź»źķź╣ļAīė SPARQLŚ╩╦„Satoshi Kume
?
Wikidataż╦ż¬ż▒żļźŪ®`ź┐ļAīėż╬SPARQLŚ╩╦„
< żŌż»żĖ >
?RDFźŪ®`ź┐źŌźŪźļż╬╗∙▒Š
?Wikidata Graph Builder200612_BioPackathon_ss
200612_BioPackathon_ssSatoshi Kume
?
This document discusses R packages related to TensorFlow and Keras, focusing on their applications in deep learning for bioinformatics, including regression, classification, and GANs. It provides a setup guide for using TensorFlow and Keras in R, along with examples of model building and transfer learning, particularly using the TensorFlow Hub with Keras. Additionally, it highlights various packages available on RStudio GitHub for TensorFlow integration and resources for working with datasets.CentOS_slide_ver1.0
CentOS_slide_ver1.0Satoshi Kume
?
This document provides instructions for installing R and using shell commands from within R on a CentOS 7 system. It explains how to install R using yum by first installing the EPEL repository and enabling it. It then shows how to use the system() function to run bash, tcsh, or other shell scripts from within R. The document also provides an example csh script and shows how to make it executable. Finally, it describes how to optionally install the GNOME desktop environment.R_note_ODE_ver1.0
R_note_ODE_ver1.0Satoshi Kume
?
Ordinary Differential Equation using R.
Note: Please check the downloaded pptx version if you want to see the GIF animations of ODE.
Kinetics_Math_ver1.0
Kinetics_Math_ver1.0Satoshi Kume
?
Ė├╬─ĄĄĮķ╔▄┴╦Ę┤ė”╦┘┬╩Ą─╩²č¦╗∙┤ĪŻ¼║ŁĖŪ┴╦▒õ┴┐Ęų└ļ╗²ĘųĘ©ĪóČ©│ŻŽĄ╩²ę╗ĮūŽ▀ąį╬óĘųĘĮ│╠╝░Ė„ųų╗»č¦Ę┤ė”└Óą═Ż©╚ń┴Ń┤╬Īóę╗ĪóČ■┤╬╝░┐╔─µĘ┤ė”Ż®Ą─╦┘┬╩ĘĮ│╠ĪŻ┤╦═ŌŻ¼╦³╗╣╠ų┬█┴╦░ļ╦źŲ┌Ą─╝Ų╦ŃŻ¼ųĖ│÷ę╗ĮūĘ┤ė”Ą─░ļ╦źŲ┌ėļ│§╩╝┼©Č╚╬▐╣žŻ¼Č°Ųõ╦¹Ę┤ė”└Óą═Ą─░ļ╦źŲ┌į“ę└└Ąė┌┼©Č╚ĪŻ╬─ĄĄ╬¬╗»č¦Ę┤ė”Č»┴”č¦╠ß╣®┴╦ŽĄ═│ąįĄ─ĘĮĘ©ėļ╣½╩ĮĪŻIgor_pro_ODE_japanese_ver2.0
Igor_pro_ODE_japanese_ver2.0 Satoshi Kume
?
Ė├╬─ĄĄĮķ╔▄┴╦│Ż╬óĘųĘĮ│╠Ż©░┐Č┘ĘĪŻ®į┌Ę┤ė”─Żą═ųąĄ─ė”ė├Ż¼░³└©ČÓĖ÷Ę┤ė”─Żą═Ą─╩²č¦▒Ē┤’║═Į┼▒Š╩Š└²ĪŻ├┐Ė÷─Żą═š╣Žų┴╦▓╗═¼Ę┤ė”╬’ų«╝õĄ─╣žŽĄęį╝░╦³├Ū╦µ╩▒╝õĄ─▒õ╗»ĪŻ╬─ĄĄ╗╣║ŁĖŪ┴╦├Ė┤┘Ę┤ė”Ą─▓╗┐╔─µ║═┐╔─µ─Żą═Ą─ĘĮ│╠╩ĮĪŻMore Related Content
More from Satoshi Kume (20)
210609 Biopackthon: BioImageDbs for ExperimentalHub (ą▐š²░µ)
210609 Biopackthon: BioImageDbs for ExperimentalHub (ą▐š²░µ)Satoshi Kume
?
BioImageDbs (for ExperimentHub)ż╬ķ_░kż╚żĮż│ż½żķ覿¾ż└źčź├ź▒®`źĖū„│╔ż╬Tipsż╦ż─żżżŲOpenlink Virtuoso v01
Openlink Virtuoso v01Satoshi Kume
?
ĪĖOpenlink VirtuosoĪ╣ż╬źżź¾ź╣ź╚®`źļż╚įOČ©ż╬╚š▒ŠšZ┘Y┴Ž
?Wikitadaź└ź¾źūż╬╚ĪĄ├
?Mac ░µ Virtuoso ż╬źżź¾ź╣ź╚®`źļż╚įOČ©
?Win░µ Virtuoso ż╬źżź¾ź╣ź╚®`źļż╚įOČ©
201209 Biopackathon 12th
201209 Biopackathon 12thSatoshi Kume
?
╗ŁŽ±äI└Ē?╗ŁŽ±ĮŌ╬÷ż╦ķvż╣żļRźčź├ź▒®`źĖż“š{¢╦żĘż┐ż╬żŪĪóęŖż─ż½ż├ż┐źčź├ź▒®`źĖż“Ż©Ä┌ż»Ū│ż»Ż®ĮBĮķż╣żļĪŻ
š{¢╦īØŽ¾: CRANĪóBioconductorĪóNeuroconductor (= GitHub/CRAN)ż╚ż½
Ś╩╦„źŁ®`ź’®`ź╔: imag(e), microscop(y) ctc
ANTsRNetż“╩╣ż├żŲĪó2D U-NET/CPU░µż“żõż├żŲż▀ż┐ĪŻ201126 Wikidata ź»źķź╣ļAīė SPARQLŚ╩╦„
201126 Wikidata ź»źķź╣ļAīė SPARQLŚ╩╦„Satoshi Kume
?
Wikidataż╦ż¬ż▒żļźŪ®`ź┐ļAīėż╬SPARQLŚ╩╦„
< żŌż»żĖ >
?RDFźŪ®`ź┐źŌźŪźļż╬╗∙▒Š
?Wikidata Graph Builder200612_BioPackathon_ss
200612_BioPackathon_ssSatoshi Kume
?
This document discusses R packages related to TensorFlow and Keras, focusing on their applications in deep learning for bioinformatics, including regression, classification, and GANs. It provides a setup guide for using TensorFlow and Keras in R, along with examples of model building and transfer learning, particularly using the TensorFlow Hub with Keras. Additionally, it highlights various packages available on RStudio GitHub for TensorFlow integration and resources for working with datasets.CentOS_slide_ver1.0
CentOS_slide_ver1.0Satoshi Kume
?
This document provides instructions for installing R and using shell commands from within R on a CentOS 7 system. It explains how to install R using yum by first installing the EPEL repository and enabling it. It then shows how to use the system() function to run bash, tcsh, or other shell scripts from within R. The document also provides an example csh script and shows how to make it executable. Finally, it describes how to optionally install the GNOME desktop environment.R_note_ODE_ver1.0
R_note_ODE_ver1.0Satoshi Kume
?
Ordinary Differential Equation using R.
Note: Please check the downloaded pptx version if you want to see the GIF animations of ODE.
Kinetics_Math_ver1.0
Kinetics_Math_ver1.0Satoshi Kume
?
Ė├╬─ĄĄĮķ╔▄┴╦Ę┤ė”╦┘┬╩Ą─╩²č¦╗∙┤ĪŻ¼║ŁĖŪ┴╦▒õ┴┐Ęų└ļ╗²ĘųĘ©ĪóČ©│ŻŽĄ╩²ę╗ĮūŽ▀ąį╬óĘųĘĮ│╠╝░Ė„ųų╗»č¦Ę┤ė”└Óą═Ż©╚ń┴Ń┤╬Īóę╗ĪóČ■┤╬╝░┐╔─µĘ┤ė”Ż®Ą─╦┘┬╩ĘĮ│╠ĪŻ┤╦═ŌŻ¼╦³╗╣╠ų┬█┴╦░ļ╦źŲ┌Ą─╝Ų╦ŃŻ¼ųĖ│÷ę╗ĮūĘ┤ė”Ą─░ļ╦źŲ┌ėļ│§╩╝┼©Č╚╬▐╣žŻ¼Č°Ųõ╦¹Ę┤ė”└Óą═Ą─░ļ╦źŲ┌į“ę└└Ąė┌┼©Č╚ĪŻ╬─ĄĄ╬¬╗»č¦Ę┤ė”Č»┴”č¦╠ß╣®┴╦ŽĄ═│ąįĄ─ĘĮĘ©ėļ╣½╩ĮĪŻIgor_pro_ODE_japanese_ver2.0
Igor_pro_ODE_japanese_ver2.0 Satoshi Kume
?
Ė├╬─ĄĄĮķ╔▄┴╦│Ż╬óĘųĘĮ│╠Ż©░┐Č┘ĘĪŻ®į┌Ę┤ė”─Żą═ųąĄ─ė”ė├Ż¼░³└©ČÓĖ÷Ę┤ė”─Żą═Ą─╩²č¦▒Ē┤’║═Į┼▒Š╩Š└²ĪŻ├┐Ė÷─Żą═š╣Žų┴╦▓╗═¼Ę┤ė”╬’ų«╝õĄ─╣žŽĄęį╝░╦³├Ū╦µ╩▒╝õĄ─▒õ╗»ĪŻ╬─ĄĄ╗╣║ŁĖŪ┴╦├Ė┤┘Ę┤ė”Ą─▓╗┐╔─µ║═┐╔─µ─Żą═Ą─ĘĮ│╠╩ĮĪŻExchange program 071128
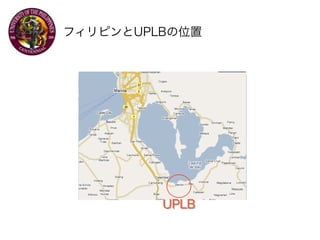
- 1. Exchange Program UNIVERSITY OF THE PHILIPPINES LOS BA?OS 20071128 Satoshi KUME
- 4. ╩┌śIż╦ż─żżżŲ Introductory Molecular Biology (Ęųūė╔·╬’č¦╚ļķT) Principles of Plant Breeding (ų▓╬’ė²ĘNż╬įŁ└Ē) Physical Education Judo (╠Õė²?╚ߥ└) ┤¾ēõļyżĘżżĪŻĪŻ
- 8. ó┘ Fż¼P░k궿╦ż╩żļĪŻ If:źżźšĪ·źżźū Co?ee:ź│źšźŻĪ·ź│źįźŻ ?fty:źšźŻźšźŲźŻĪ·źįźŻźūźŲźŻ ó┌ LEż¼ELż╦ż╩żļĪŻ singleĪ·singel : źĘź¾ź▓źļ bicycleĪ·bicycel : źąźżźĘź▒źļ ó█ ThżŽź┐ĪóRżŽźļ░kę¶ thirty: ź┐źļźŲźŻ źšźŻźĻźįź¾ėóšZż╦æTżņżļĪŻĪŻĪŻ ÅĻė├ Farmer :?▐r╝ę źčźļź▐źļ
- 9. źšźŻźĻźįź¾żŪż╬ėóšZż╬Ąž╬╗ ╗∙▒ŠĄ─ż╦ź┐ź¼źĒź░šZż“įÆż╣ĪŻ źšźŻźĻźįź¾╚╦ż¼źšźŻźĻźįź¾żŪėóšZż¼įÆż╗żļ Ą▒╚╗ĪŻĪŻ
- 11. ═¼Šė╚╦ż┐ż┴
- 12. Maraming salamat po... Thank you very much...
