Genomics seminar
- 1. Genomics Presented by: Samaneh.Rasoulinejad Fall 2013 - 2014
- 2. What is Genomics? âĒ âĒ Genomcis is the study of all genes present in an organism In 1986 mouse geneticist Thomas Roderick used Genomics for âmapping, sequencing and characterizing genomesâ
- 3. Introduction âĒ Genomics built on recombinant-DNA technology (developed since early 1970s) âĒ Thorough understanding of recombinant-DNA techniques âĒ Prerequisite for understanding genomics technologies âĒ Differences between genomics and recombinant-DNA technology âĒ Genomics is high throughput approaches to allow more analyses in parallel âĒ Genomics is dependent on computational analysis due to larger data sets
- 4. ïķSequence the entire genome by cutting it into small, manageable pieces (fragments) ïķAssemble the entire genome from the pieces (fragments) ïķMake sense of the genome ïķUnderstand how gene expression takes place? ïķHow life processes are networked? ïķUnderstand life??
- 5. Technical Foundations of ïGenomic and cDNA libraries Genomics ïDNA Hybridization and Northern blots ïSubcloning in vectors ïRestriction-enzyme mapping ïDNA sequencing ïPCR amplification Genomics and Medicine
- 6. What we hope to gain from genomics -Drug, diagnostics, and prognostics development - Genotyping to predict patient susceptibility to disease - Personalized healthcare based on an individualâs genomic features genome decision support systems genotype molecular profile patient history knowledge base drugs diagnostics prognostics health
- 7. Over 1,000 disease genes were characterized by 2000
- 8. How to make a genomic library ori total genomic DNA ampR genomic DNA restriction enzyme anneal and ligate ori ampR ori plasmid (black) ampR ori ampR ori ampR same restriction enzyme transform E. coli; select for Amp resistance
- 9. selected colonies tissue or cell membrane mRNA polyA stationary support polyT Radioactive probe plasmid E. Coli bacteria hybridization X-ray film cDNA library Clone 1 2 3 4 5
- 11. Microarrays âĒ Basis of microarrays for determining gene expression âĒ Process by which complementary strands find each other âĒ AâT and CâG base pairing âĒ speed and fidelity: dependent on temperature, salt, sequence, and concentration (High temp and low salt)
- 12. âĒ âĒ âĒ âĒ Microarrays permit the simultaneous analysis of the RNA expression of thousands of genes. For fully sequenced genomes, microarrays can be used to analyze the expression of every gene. Prior to the introduction of microarrays, RNA abundance was usually analyzed through hybridization to RNA bound to filters. These Northern blots normally had no more than 20â40 lanes, and no more than three probes could be used simultaneously. In contrast, microarrays can interrogate 30,000 genes at the same time, vastly increasing our ability to analyze RNA expression.
- 13. Northern blot and microarray 0 2 5 6 7 hrs 0 2 5 6 7 9 11 hrs DMC1 â DMC1 â SPS1 â DIT1 â SPS1 â SPS100 â 0 2 5 6 7 9 11 hrs DIT1 â SPS100 â fold repressed fold induced >20 10x 3x | 3x 10x >20 1:1 Identify genes whose expression was induced during sporulation in yeast
- 14. Cross-hybridization âĒ âĒ âĒ âĒ Hybridization to a related, but not identical, sequence = cross-hybridization Example: A probe from one member of a gene family is likely to hybridize to all other members Problem in microarrays, particularly cDNA arrays Oligonucleotide arrays prescreened to eliminate sequences likely to cross-hybridize
- 15. Improved disease diagnostics from genomics âĒ Microarray analysis of gene expression from four different types of tumors
- 16. Microarrays and cancer âĒ âĒ Histology not always effective tool for prognosis and diagnosis Microarrays distinguish cancerous tissues on the basis of a gene expression profile âĒ Use in diagnosis (presence) âĒ âĒ Example: characterizing acute lymphoblastic leukemia. Also breast cancer. Use in prognosis âĒ Example: assessing the likelihood of metastasis in medulloblastoma (brain tumor in children)
- 17. Microarrays in the prognosis of metastasis (childhood cancer: medulloblastoma) âĒ âĒ âĒ âĒ Identified 85 genes with different levels of expression in metastatic (M+) and nonmetastatic tumors (M_) 59 up and 26 down 72% accuracy in predicting metastasis Identified genes induced in metastasis âĒ Could serve as potential drug targets for in vitro experiments âĒ platelet derived growth factor receptor alpha (PDGFRÎą). Antibodies prevent migration. Mâ M+ green = down regulated red = up regulated
- 18. Cancer genome projects One in three people will suffer from cancer in his or her lifetime. Cancer Genome Anatomy Project (CGAP) Established 1997 by National Cancer Institute (USA) Specializes in EST sequencing Human Cancer Genome Project (HCGP) Established 1999 by Brazilian research groups Cancer Genome Project (CGP) Established 2000 by Wellcome Trust and Sanger Institute (United Kingdom) Specializes in genomic mutations leading to cancer Funding: $15 million to $60 million
- 19. Classes of microarrays âĒ Custom/spotted/two-color microarrays (cDNAs, BACs) âĒ High-density oligonucleotide arrays (GeneChip, Affymetrix) âĒ Long oligonucleotide microarrays - Agilent (25-60 bases) - Illumina (50 bases) - Nimblegen (50-75 bases)
- 20. Affymetrix oligonucleotide arrays The array elements are a series of 25mer oligos designed from known sequence and synthesized Directly on the surface The entire array is formed by >500,000 cells, each containing a different oligo
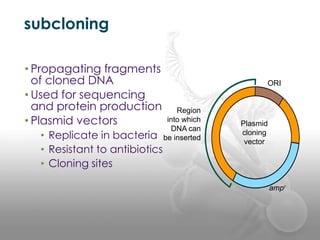
- 21. subcloning âĒ Propagating fragments of cloned DNA âĒ Used for sequencing and protein production âĒ Plasmid vectors âĒ Replicate in bacteria âĒ Resistant to antibiotics âĒ Cloning sites ORI Region into which DNA can be inserted Plasmid cloning vector ampr
- 22. Subcloning: vector and fragment âĒ Vector and fragment to be DNA fragment inserted must have compatible ends âĒ Sticky ends anneal âĒ Enzyme ligase makes covalent cloning vector bond between vector and fragment âĒ Use of recombination instead of restriction sites recombinant plasmid restriction enzymes
- 23. Recombination cloning âĒ âĒ âĒ âĒ Uses site-specific recombination for subcloning DNA fragment flanked by recombination sites Add recombinase âClonaseÂŪâ Moves fragment from one vector to another
- 24. DNA sequencing âĒ Most current sequencing projects use the chain termination method âĒ âĒ Based on action of DNA polymerase âĒ âĒ Also known as Sanger sequencing, after its inventor, Fredrick Sanger Adds nucleotides to complementary strand Requires template DNA and primer
- 26. Chain-termination sequencing âĒ Dideoxynucleotides stop synthesis âĒ Chain terminators Included in amounts so as to terminate every time the base appears in the template âĒ Use four reactions âĒ âĒ One for each base: A,C,G, and T
- 28. Sequence detection To detect products of sequencing reaction Include labeled nucleotides Formerly, radioactive labels were used Now fluorescent labels Use different fluorescent tag for each nucleotide Can run all four reactions in same lane
- 29. Pyrosequencing âĒ âĒ âĒ based on the sequencing by synthesis principle. it relies on the detection of pyrophosphate release on nucleotide incorporation The technique was developed by Mostafa Ronaghi and PÃĨl NyrÃĐn at the Royal Institute of Technology in Stockholm in 1996
- 32. PCRs âĒ âĒ âĒ âĒ âĒ âĒ âĒ âĒ âĒ âĒ Colony PCR Helicase PCR Hot- start In situ Intersequence specific Inverse Multiplex Quantitative Touch down ......
- 33. Resources âĒ Bioinformatics, Genomics, and Proteomics (Ann Finney Batiza, Ph.D.) âĒ SciencePages âĒ Functional Genomics (Michael Kaufmann and Claudia Klinger Private Universitt, Witten/Herdecke gGmbH, Witten, Germany) Introduction to Genomics by Arthur M. Lesk, 2007, Oxford University Press
- 34. Thank you