Integrate JBrowse REST API Framework with Adama Federation Architecture
- 1. araport.org@araport Integrate JBrowse REST API with ADAMA Vivek Krishnakumar 12/03/2015
- 2. araport.org@araport Outline ŌĆó JBrowse REST API spec ŌĆó ADAMA endpoints ŌĆó Mapping JBrowse to ADAMA endpoints ŌĆó Extending JBrowse REST feature store ŌĆó Handling Auth in JBrowse 1.x ŌĆó Use Case ŌĆō Integrate Chromatin States data into JBrowse via ADAMA ŌĆó Summary ŌĆó Challenges
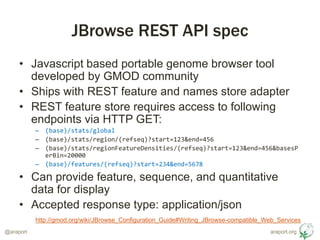
- 3. araport.org@araport JBrowse REST API spec ŌĆó Javascript based portable genome browser tool developed by GMOD community ŌĆó Ships with REST feature and names store adapter ŌĆó REST feature store requires access to following endpoints via HTTP GET: ŌĆō (base)/stats/global ŌĆō (base)/stats/region/(refseq)?start=123&end=456 ŌĆō (base)/stats/regionFeatureDensities/(refseq)?start=123&end=456&basesP erBin=20000 ŌĆō (base)/features/(refseq)?start=234&end=5678 ŌĆó Can provide feature, sequence, and quantitative data for display ŌĆó Accepted response type: application/json http://gmod.org/wiki/JBrowse_Configuration_Guide#Writing_JBrowse-compatible_Web_Services
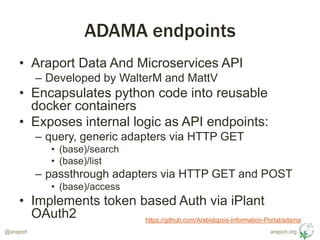
- 4. araport.org@araport ADAMA endpoints ŌĆó Araport Data And Microservices API ŌĆō Developed by WalterM and MattV ŌĆó Encapsulates python code into reusable docker containers ŌĆó Exposes internal logic as API endpoints: ŌĆō query, generic adapters via HTTP GET ŌĆó (base)/search ŌĆó (base)/list ŌĆō passthrough adapters via HTTP GET and POST ŌĆó (base)/access ŌĆó Implements token based Auth via iPlant OAuth2 https://github.com/Arabidopsis-Information-Portal/adama
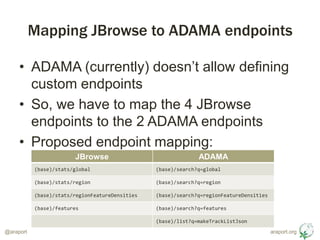
- 5. araport.org@araport Mapping JBrowse to ADAMA endpoints ŌĆó ADAMA (currently) doesnŌĆÖt allow defining custom endpoints ŌĆó So, we have to map the 4 JBrowse endpoints to the 2 ADAMA endpoints ŌĆó Proposed endpoint mapping: JBrowse ADAMA (base)/stats/global (base)/search?q=global (base)/stats/region (base)/search?q=region (base)/stats/regionFeatureDensities (base)/search?q=regionFeatureDensities (base)/features (base)/search?q=features (base)/list?q=makeTrackListJson
- 6. araport.org@araport Extending JBrowse REST feature store ŌĆó JBrowse/Store/SeqFeature/REST handles requests from the browser to the configured API service ŌĆō Understands the 4 pre-defined endpoints ŌĆó REST feature store extended based on proposed endpoint mapping ŌĆó Utilize JBrowse Plugin architecture to develop Araport plugin: Araport/Store/SeqFeature/REST http://gmod.org/wiki/JBrowse_Configuration_Guide#Writing_JBrowse_Plugins
- 7. araport.org@araport Handling Auth in JBrowse 1.x ŌĆó ADAMA (currently) requires all requests to be accompanied by an Authorization Bearer $TOKEN ŌĆó JBrowse 1.x does not support OAuth2 implicit flow, thus cannot generate tokens ŌĆó Solution: ŌĆō OAuth2 $TOKEN created with unlimited validity (-1) ŌĆó $TOKEN generated by Araport anonymous user ŌĆó $TOKEN scope limited to ADAMA services ŌĆō Araport/Store/SeqFeature/RESTAuth hard codes $TOKEN ŌĆō Araport/Store/SeqFeature/REST inherits from RESTAuth
- 8. araport.org@araport Use Case Integrate Chromatin States data into JBrowse via ADAMA ŌĆó Published dataset representing a landscape of Arabidopsis thaliana chromatin states using 16 features, including DNA sequence, CG methylation, histone variants, and modifications ŌĆó Data represents chromosomal regions (start/stop coordinates) corresponding with 9 distinct chromatin states ŌĆó Published with manuscript as supplementary dataset (in XLS format)Sequeira-Mendes, J., Arag├╝ez, I., Peir├│, R., Mendez-Giraldez, R., Zhang, X., Jacobsen, S. E., ... & Gutierrez, C. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. The Plant Cell, 26(6), 2351-2366.
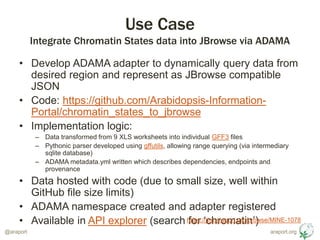
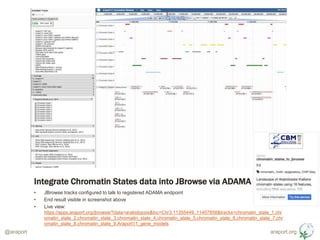
- 9. araport.org@araport ŌĆó Develop ADAMA adapter to dynamically query data from desired region and represent as JBrowse compatible JSON ŌĆó Code: https://github.com/Arabidopsis-Information- Portal/chromatin_states_to_jbrowse ŌĆó Implementation logic: ŌĆō Data transformed from 9 XLS worksheets into individual GFF3 files ŌĆō Pythonic parser developed using gffutils, allowing range querying (via intermediary sqlite database) ŌĆō ADAMA metadata.yml written which describes dependencies, endpoints and provenance ŌĆó Data hosted with code (due to small size, well within GitHub file size limits) ŌĆó ADAMA namespace created and adapter registered ŌĆó Available in API explorer (search for ŌĆśchromatinŌĆÖ) Use Case Integrate Chromatin States data into JBrowse via ADAMA https://jira.araport.org/browse/MINE-1078
- 10. araport.org@araport Integrate Chromatin States data into JBrowse via ADAMA ŌĆó JBrowse tracks configured to talk to registered ADAMA endpoint ŌĆó End result visible in screenshot above ŌĆó Live view: https://apps.araport.org/jbrowse/?data=arabidopsis&loc=Chr3:11355449..11407858&tracks=chromatin_state_1,chr omatin_state_2,chromatin_state_3,chromatin_state_4,chromatin_state_5,chromatin_state_6,chromatin_state_7,chr omatin_state_8,chromatin_state_9,Araport11_gene_models
- 11. araport.org@araport Summary ŌĆó Extended JBrowse to integrate with ADAMA endpoints ŌĆó Exposed a published community dataset via ADAMA as an API ŌĆó Exposed track configuration via ADAMA API ŌĆó Visualized dataset in JBrowse via ADAMA API
- 12. araport.org@araport Challenges ŌĆó Making web service calls to ADAMA requires Authorization $TOKEN ŌĆó JBrowse needs to hard code the Authorization $TOKEN ŌĆó Track configuration needs to be manually added to JBrowse ŌĆó Scaling to larger datasets not feasible via GitHub route
- 13. araport.org@araport Acknowledgements ŌĆó JCVI Araport Team ŌĆō Chia-Yi Cheng ŌĆō Agnes Chan ŌĆō Chris Town ŌĆó TACC Araport Team ŌĆō Walter Moreira ŌĆō Matthew Vaughn ŌĆó Prof. Crisanto Gutierrez (CBMSO)