Microbial phylogeny
Download as DOCX, PDF7 likes9,074 views
Microbial Phyllogeny.and Classification. Phyllogenetic Tree , Domain and determining technique.
1 of 5
Downloaded 44 times
![Microbial Phylogeny: Classification of species in
superior taxa and construction of phylogenetic trees based
on evolutionary relationships.
It is bringing order to the diverse variety of organisms present
in nature.
So there are two general ways the classification can
be constructed. First one is based on the
morphological characters (phenetic classification) and
second is based on evolutionary relationship
(phylogenetic classification)
Phenetic classification – It is based on the mutual
similarity of their phenotypic characteristics. It does not
provide any information about phylogenetic relations.
Phylogenetic classification- These systems are based on
evolutionary relationships rather than external appearance
.The term phylogeny [Greek phylon, tribe or race,
and genesis, generation or origin] refers to the evolutionary
development of a species. It is based on the direct comparison
of genetic materials and gene product.](https://image.slidesharecdn.com/microbialphylogeny-170814080323/85/Microbial-phylogeny-1-320.jpg)




Recommended
Microbial taxonomy and classification system



Microbial taxonomy and classification systemSakshi Saxena
Ěý
- Taxonomy is the science of describing, naming, and classifying organisms. It provides understanding of biodiversity which is important for conservation and sustainability.
- Aristotle was the first to attempt classifying organisms by type and introduce binomial nomenclature. Later systems were proposed by Linnaeus, Whittaker, and Woese based on new understandings of cell structure, genetics, and evolution.
- Different classification systems include artificial, natural, phylogenetic, polyphasic, and numerical taxonomy which use varying characteristics and methodologies.Lamda phage



Lamda phageMinhaz Ahmed
Ěý
Lambda phage is a bacteriophage that infects E. coli bacteria. It has two life cycles: a lytic cycle and a lysogenic cycle. In the lytic cycle, the phage genome is transferred into the bacterial cell where it replicates and causes the bacterial cell to burst, releasing new phage particles. In the lysogenic cycle, the phage genome integrates into the bacterial chromosome and replicates with the host DNA without killing the cell. The phage can switch between these two cycles depending on environmental conditions inside the infected bacterial cell.Biodegradation of xenobiotics



Biodegradation of xenobioticsgaurav raja
Ěý
The document summarizes biodegradation of xenobiotic compounds, specifically petroleum hydrocarbons and pesticides. It discusses how various microorganisms can degrade these compounds through aerobic and anaerobic pathways. Key points include how bacteria and enzymes are able to break down petroleum, degrade pesticides, and transform toxic contaminants into less hazardous substances through microbial metabolic pathways and catabolic reactions. Recent research is also cited that studied biodegradation of crude oil by bacterial consortium in the marine environment.Bacteriophage T4 and Bacteriophage lambda 



Bacteriophage T4 and Bacteriophage lambda HARINATHA REDDY ASWARTHAGARI
Ěý
This document discusses two types of bacteriophages - T4 and Lambda phages. It describes their structure, genetic material, life cycles and infection processes. T4 phage infects E. coli and has a long helical tail and base plate with spikes that attach to host cells. Lambda phage also infects E. coli and can undergo lysogenic or lytic cycles. It has genes that determine which cycle occurs based on host cell conditions. The document provides detailed information on the structure, replication, and infection mechanisms of these two important bacteriophages.Microbial taxonomy



Microbial taxonomyTamil Silambarasan
Ěý
M. Sc., Microbiology Study Material (Semester-I, General Microbiology)
Department of Microbiology, School of Bioscience,
Periyar University, Salem.Prokaryotic genome organization



Prokaryotic genome organizationmanojsiddartha bolthajira
Ěý
Prokaryotic genomes are typically organized as single circular chromosomes that are condensed into a nucleoid region within the cell. DNA supercoiling, which involves the over- or under-winding of DNA strands, facilitates compaction of prokaryotic genomes and enables DNA metabolism. Topoisomerases regulate DNA supercoiling by introducing temporary breaks in DNA strands, allowing strands to pass through one another and relieve torsional stress that builds during processes like transcription and replication. The two major types of topoisomerase are types I and II, which introduce single-strand or double-strand breaks, respectively, in regulating supercoiling levels.Plant and animal viruse



Plant and animal viruseNikita Dewangan
Ěý
This presentation about virus - virus structure, effect of virus on animal and plant pathogenicity, type of virus.Animal viruses



Animal virusesMonika K Raut
Ěý
Animal viruses are self replicating, intracellular parasites that completely rely on host animal cell for reproduction. They use the host's cellular components to replicate, then leaves the host cell to infect other cells.
Baculoviruses as biocontrol agents



Baculoviruses as biocontrol agentsSonia John
Ěý
Baculoviruses are viruses that can infect and kill many invertebrate organisms, including insects. They are usually small and contain double-stranded DNA. Baculoviruses can remain dormant in the environment for long periods before infecting insects. Most baculoviruses must be eaten by the host insect to cause infection. Genetic engineering has enhanced baculoviruses for use as biological insecticides by introducing genes that increase the speed of killing insects. Introduced genes include genes for Bt toxin, scorpion neurotoxin, and other toxins. Baculoviruses are good candidates for species-specific pest control due to their ability to persist in the environment and multiply rapidly withinNomenclature, classification and identification of bacteria by Anil Shrestha



Nomenclature, classification and identification of bacteria by Anil ShresthaAnil Shrestha
Ěý
This document discusses various terminologies and techniques used in microbial taxonomy and classification. It defines key terms like nomenclature, classification, and identification. It also describes different classical methods of classification like morphology, physiology, biochemistry, and genetics. Additionally, it outlines modern molecular methods such as nucleic acid sequencing, genomic fingerprinting, and amino acid sequencing that provide stronger evidence for microbial evolution and phylogeny. The document emphasizes that a polyphasic approach integrating both classical and molecular data provides the most accurate microbial taxonomy.Numerical taxonomy



Numerical taxonomyKARTHIK REDDY C A
Ěý
Numerical taxonomy is a classification system that uses numerical methods to group taxonomic units based on their character states. It was developed in the 1950s alongside multivariate analysis and computer development. Numerical taxonomy aims to define homogeneous clusters of organisms by integrating different data types through statistical analysis. This results in a phenogram or dendrogram that illustrates the relationships between taxa based on their similarities. Numerical taxonomy is useful for studying relationships between bacteria and other microorganisms.Bergey’s manual of bacterial classification



Bergey’s manual of bacterial classificationnj1992
Ěý
Bergey’s Manual is a key reference work for classifying and identifying prokaryotes. It consists of two parts: Bergey’s Manual of Determinative Bacteriology, which provides identification schemes based on morphology and biochemical tests; and Bergey’s Manual of Systematic Bacteriology, which provides phylogenetic classification based on rRNA sequencing. The first edition in 1923 established bacterial classification for identification. Current editions are based on phylogenetic studies and distribute pathogenic bacteria throughout volumes organized by phylogenetic relationships rather than clinical importance.Ti plasmid



Ti plasmidArunima Sur
Ěý
The document summarizes a seminar on the Ti plasmid. It discusses that the Ti plasmid is found in Agrobacterium tumefaciens and is responsible for crown gall tumor formation in plants. It describes the organization and structure of the Ti plasmid, including the T-DNA region flanked by borders that is transferred to plant cells. Two common vector systems used for plant transformation, the cointegrate vector and binary vector, are explained. The cointegrate vector involves integration of an intermediate vector with the Ti plasmid, while the binary vector separates the plasmid and virulence genes. Finally, the general process of Agrobacterium-mediated plant transformation is outlined.Morphology and cell structure of Archea



Morphology and cell structure of ArcheaSanjay Nagal
Ěý
Archea are primitive organisms of this world. They are different from bacteria.They are termed as archea. Genome organization in prokaryotes



Genome organization in prokaryotesSangeeta Das
Ěý
This ppt describes about the various stages as well as the enzymes involved in the process of genome organization in prokaryotes.Operon 



Operon sijiskariah
Ěý
This document provides an overview of gene regulation in prokaryotes using the lac operon in E. coli as an example. It explains that genes are regulated to control which proteins are expressed at different times. The lac operon consists of structural genes that encode enzymes for lactose metabolism, as well as a regulatory gene that produces a repressor protein. In the absence of the lactose inducer, the repressor binds to the operator region and prevents transcription. When lactose is present, it binds to the repressor and causes a conformational change that prevents it from binding to the operator, allowing transcription.Lectut btn-202-ppt-l4. bacteriophage lambda and m13 vectors (1)



Lectut btn-202-ppt-l4. bacteriophage lambda and m13 vectors (1)Rishabh Jain
Ěý
This document describes the bacteriophages λ and M13, which are commonly used as cloning vectors. λ phage is a temperate phage that infects E. coli and has a double-stranded linear DNA genome. Its genome is organized into regions that encode proteins for the phage head, tail, and lysogeny/lysis functions. M13 is a filamentous phage with a single-stranded circular genome. Both phages can be modified and used to insert and replicate foreign DNA fragments in E. coli for cloning purposes.Abzymes



Abzymesjeeva raj
Ěý
Abzymes, also known as catalytic antibodies, are monoclonal antibodies that exhibit enzymatic activity. They are able to bind to transition states of enzyme-catalyzed reactions with high specificity and affinity, stabilizing the transition state and increasing reaction rates. Abzymes can be artificially produced by immunizing animals with transition state analogs of reactions. They have potential applications in drug development, cancer treatment, and developing therapies for viral infections like HIV. Researchers have engineered an abzyme that can degrade an essential region of the HIV envelope protein, rendering the virus unable to infect cells.Development of drosophila



Development of drosophilaKAUSHAL SAHU
Ěý
Introduction
About Drosophila
Genome of Drosophila
Life cycle
Differentiation
Development of Drosophila
* Embryonic development
* Dorsal -ventral and
* Anterior posterior development
* Body segmentation
* Homeotic gene
Conclusion
Reference
Bacterial growth : Diauxic growth,Synchronous growth and continuous growth



Bacterial growth : Diauxic growth,Synchronous growth and continuous growthSivasangari Shanmugam
Ěý
It contain bacterial growth,growth rate and how do it grow?
and,I include Types of continuous growth and their advantages,disadvantages.Measurement of microbial growth



Measurement of microbial growthNOOR ARSHIA
Ěý
Direct methods of measurement of microbial growth includes various methods of enumeration of both viable and non viable cell also includes growth curve. Helpful for UG and PG programs of microbiology
THE ARCHAEA



THE ARCHAEAMariya Raju
Ěý
This document provides information about the six kingdoms of life and the domains of Archaea and Bacteria. It discusses how biologists classify organisms and some of the key distinguishing characteristics, such as cell structure, nutrition, and reproduction. Specifically, it outlines that Archaea and Bacteria are both prokaryotic domains. Archaea are found in extreme environments and have unique cell wall composition and membrane lipids. Bacteria are more ubiquitous and also have rigid cell walls containing peptidoglycan. Many bacteria can be pathogenic if they enter the body and use tissues as a food source or secrete toxins.Bergey's classification presentation



Bergey's classification presentationBIDISHA MANDAL
Ěý
Bergey's Manual and it's classification. A brief concised presentation prepared for taking seminar and classes.
Volume II (Edition 2) described more in detail.Photosynthesis in bacteria and its types



Photosynthesis in bacteria and its typesSaajida Sultaana
Ěý
This document summarizes different types of bacterial photosynthesis. It describes oxygenic photosynthesis, which is similar to plant photosynthesis and results in oxygen production. This type is found in cyanobacteria. It also describes anoxygenic photosynthesis, which does not produce oxygen. This type is found in purple and green sulfur bacteria, and involves using hydrogen sulfide or hydrogen as electron donors instead of water. The document provides examples of different bacteria that use anoxygenic photosynthesis and details their characteristics and where they are typically found.Biodegradation of pesticides



Biodegradation of pesticidesaachal jain
Ěý
1. Biodegradation is the process by which microorganisms like bacteria and fungi break down pesticides into non-toxic substances.
2. Common pesticides that are biodegraded include the soil fumigant methyl bromide, the herbicide dalapon, and the fungicide chloroneb.
3. For effective biodegradation, organisms must be able to degrade the pesticide, the pesticide must be bioavailable, and soil conditions must support microbial growth. Strategies to enhance biodegradation include biostimulation, bioventing, and bioaugmentation.Mycoplasma



MycoplasmaSchool of Studies in Microbiology
Ěý
This document summarizes a seminar presentation on Mycoplasma. Key points include:
- Mycoplasma are the smallest bacteria capable of autonomous growth. They lack cell walls and are sensitive to osmotic shock and antibiotics like tetracyclines.
- They were first discovered in 1843 and isolated from cattle pleural fluid. Techniques for culturing mycoplasma were developed in the late 19th century.
- Mycoplasma can take different shapes and sizes. They reproduce through binary fission or budding.
- They are classified as facultative anaerobes in the class Mollicutes. Some require sterols for growth while others do not. genetically modified organisms in the field of bio-remediation



genetically modified organisms in the field of bio-remediationswayam prakas nanda
Ěý
genetically modified organisms used for remediation of heavy metals, radioactive waste for bio-remediation Cot curve 



Cot curve EmaSushan
Ěý
Cot value and Cot Curve analysis is a technique for measuring DNA complexity based on renaturation kinetics. DNA is denatured and allowed to reanneal, with larger DNA taking longer. Cot value accounts for DNA concentration, time, and buffer effects, representing repetitive sequences - lower Cot means more repeats. Examples show bacteria have nearly all single-copy DNA, while mouse has varying proportions of single-copy, middle repetitive, and highly repetitive sequences. Cot curve analysis provides information on genome size, complexity, and proportions of sequence types.Ap Chapter 26 Evolutionary History Of Biological Diversity



Ap Chapter 26 Evolutionary History Of Biological Diversitysmithbio
Ěý
- Phylogeny investigates the evolutionary history and relationships between species through analysis of fossil, molecular, and genetic data. Systematists use phylogenetic trees to depict these relationships.
- A phylogenetic tree represents hypotheses about evolutionary divergence. Each branch point indicates where two lineages diverged from a common ancestor. Tracing shared derived characteristics helps systematists determine which groupings form monophyletic clades.
- Molecular systematics uses comparisons of DNA and other molecules to infer relatedness and reconstruct evolutionary history over hundreds of millions of years, helping to extend phylogenies beyond what can be learned from fossils alone. Molecular clocks and neutral theory aim to date evolutionary events but have limitations.SYSTEMATICS.pdf



SYSTEMATICS.pdfNopiImrael
Ěý
1. The document discusses various methods used to classify living things, including morphology, developmental characteristics, evolutionary descent, anatomical resemblance, and molecular information such as DNA sequences.
2. Taxonomy is the scientific classification of organisms and systematics is the study of the evolutionary relationships between organisms.
3. DNA sequencing and comparing DNA strands between organisms has provided new insights into evolutionary relationships, with more similar DNA indicating closer relation.More Related Content
What's hot (20)
Baculoviruses as biocontrol agents



Baculoviruses as biocontrol agentsSonia John
Ěý
Baculoviruses are viruses that can infect and kill many invertebrate organisms, including insects. They are usually small and contain double-stranded DNA. Baculoviruses can remain dormant in the environment for long periods before infecting insects. Most baculoviruses must be eaten by the host insect to cause infection. Genetic engineering has enhanced baculoviruses for use as biological insecticides by introducing genes that increase the speed of killing insects. Introduced genes include genes for Bt toxin, scorpion neurotoxin, and other toxins. Baculoviruses are good candidates for species-specific pest control due to their ability to persist in the environment and multiply rapidly withinNomenclature, classification and identification of bacteria by Anil Shrestha



Nomenclature, classification and identification of bacteria by Anil ShresthaAnil Shrestha
Ěý
This document discusses various terminologies and techniques used in microbial taxonomy and classification. It defines key terms like nomenclature, classification, and identification. It also describes different classical methods of classification like morphology, physiology, biochemistry, and genetics. Additionally, it outlines modern molecular methods such as nucleic acid sequencing, genomic fingerprinting, and amino acid sequencing that provide stronger evidence for microbial evolution and phylogeny. The document emphasizes that a polyphasic approach integrating both classical and molecular data provides the most accurate microbial taxonomy.Numerical taxonomy



Numerical taxonomyKARTHIK REDDY C A
Ěý
Numerical taxonomy is a classification system that uses numerical methods to group taxonomic units based on their character states. It was developed in the 1950s alongside multivariate analysis and computer development. Numerical taxonomy aims to define homogeneous clusters of organisms by integrating different data types through statistical analysis. This results in a phenogram or dendrogram that illustrates the relationships between taxa based on their similarities. Numerical taxonomy is useful for studying relationships between bacteria and other microorganisms.Bergey’s manual of bacterial classification



Bergey’s manual of bacterial classificationnj1992
Ěý
Bergey’s Manual is a key reference work for classifying and identifying prokaryotes. It consists of two parts: Bergey’s Manual of Determinative Bacteriology, which provides identification schemes based on morphology and biochemical tests; and Bergey’s Manual of Systematic Bacteriology, which provides phylogenetic classification based on rRNA sequencing. The first edition in 1923 established bacterial classification for identification. Current editions are based on phylogenetic studies and distribute pathogenic bacteria throughout volumes organized by phylogenetic relationships rather than clinical importance.Ti plasmid



Ti plasmidArunima Sur
Ěý
The document summarizes a seminar on the Ti plasmid. It discusses that the Ti plasmid is found in Agrobacterium tumefaciens and is responsible for crown gall tumor formation in plants. It describes the organization and structure of the Ti plasmid, including the T-DNA region flanked by borders that is transferred to plant cells. Two common vector systems used for plant transformation, the cointegrate vector and binary vector, are explained. The cointegrate vector involves integration of an intermediate vector with the Ti plasmid, while the binary vector separates the plasmid and virulence genes. Finally, the general process of Agrobacterium-mediated plant transformation is outlined.Morphology and cell structure of Archea



Morphology and cell structure of ArcheaSanjay Nagal
Ěý
Archea are primitive organisms of this world. They are different from bacteria.They are termed as archea. Genome organization in prokaryotes



Genome organization in prokaryotesSangeeta Das
Ěý
This ppt describes about the various stages as well as the enzymes involved in the process of genome organization in prokaryotes.Operon 



Operon sijiskariah
Ěý
This document provides an overview of gene regulation in prokaryotes using the lac operon in E. coli as an example. It explains that genes are regulated to control which proteins are expressed at different times. The lac operon consists of structural genes that encode enzymes for lactose metabolism, as well as a regulatory gene that produces a repressor protein. In the absence of the lactose inducer, the repressor binds to the operator region and prevents transcription. When lactose is present, it binds to the repressor and causes a conformational change that prevents it from binding to the operator, allowing transcription.Lectut btn-202-ppt-l4. bacteriophage lambda and m13 vectors (1)



Lectut btn-202-ppt-l4. bacteriophage lambda and m13 vectors (1)Rishabh Jain
Ěý
This document describes the bacteriophages λ and M13, which are commonly used as cloning vectors. λ phage is a temperate phage that infects E. coli and has a double-stranded linear DNA genome. Its genome is organized into regions that encode proteins for the phage head, tail, and lysogeny/lysis functions. M13 is a filamentous phage with a single-stranded circular genome. Both phages can be modified and used to insert and replicate foreign DNA fragments in E. coli for cloning purposes.Abzymes



Abzymesjeeva raj
Ěý
Abzymes, also known as catalytic antibodies, are monoclonal antibodies that exhibit enzymatic activity. They are able to bind to transition states of enzyme-catalyzed reactions with high specificity and affinity, stabilizing the transition state and increasing reaction rates. Abzymes can be artificially produced by immunizing animals with transition state analogs of reactions. They have potential applications in drug development, cancer treatment, and developing therapies for viral infections like HIV. Researchers have engineered an abzyme that can degrade an essential region of the HIV envelope protein, rendering the virus unable to infect cells.Development of drosophila



Development of drosophilaKAUSHAL SAHU
Ěý
Introduction
About Drosophila
Genome of Drosophila
Life cycle
Differentiation
Development of Drosophila
* Embryonic development
* Dorsal -ventral and
* Anterior posterior development
* Body segmentation
* Homeotic gene
Conclusion
Reference
Bacterial growth : Diauxic growth,Synchronous growth and continuous growth



Bacterial growth : Diauxic growth,Synchronous growth and continuous growthSivasangari Shanmugam
Ěý
It contain bacterial growth,growth rate and how do it grow?
and,I include Types of continuous growth and their advantages,disadvantages.Measurement of microbial growth



Measurement of microbial growthNOOR ARSHIA
Ěý
Direct methods of measurement of microbial growth includes various methods of enumeration of both viable and non viable cell also includes growth curve. Helpful for UG and PG programs of microbiology
THE ARCHAEA



THE ARCHAEAMariya Raju
Ěý
This document provides information about the six kingdoms of life and the domains of Archaea and Bacteria. It discusses how biologists classify organisms and some of the key distinguishing characteristics, such as cell structure, nutrition, and reproduction. Specifically, it outlines that Archaea and Bacteria are both prokaryotic domains. Archaea are found in extreme environments and have unique cell wall composition and membrane lipids. Bacteria are more ubiquitous and also have rigid cell walls containing peptidoglycan. Many bacteria can be pathogenic if they enter the body and use tissues as a food source or secrete toxins.Bergey's classification presentation



Bergey's classification presentationBIDISHA MANDAL
Ěý
Bergey's Manual and it's classification. A brief concised presentation prepared for taking seminar and classes.
Volume II (Edition 2) described more in detail.Photosynthesis in bacteria and its types



Photosynthesis in bacteria and its typesSaajida Sultaana
Ěý
This document summarizes different types of bacterial photosynthesis. It describes oxygenic photosynthesis, which is similar to plant photosynthesis and results in oxygen production. This type is found in cyanobacteria. It also describes anoxygenic photosynthesis, which does not produce oxygen. This type is found in purple and green sulfur bacteria, and involves using hydrogen sulfide or hydrogen as electron donors instead of water. The document provides examples of different bacteria that use anoxygenic photosynthesis and details their characteristics and where they are typically found.Biodegradation of pesticides



Biodegradation of pesticidesaachal jain
Ěý
1. Biodegradation is the process by which microorganisms like bacteria and fungi break down pesticides into non-toxic substances.
2. Common pesticides that are biodegraded include the soil fumigant methyl bromide, the herbicide dalapon, and the fungicide chloroneb.
3. For effective biodegradation, organisms must be able to degrade the pesticide, the pesticide must be bioavailable, and soil conditions must support microbial growth. Strategies to enhance biodegradation include biostimulation, bioventing, and bioaugmentation.Mycoplasma



MycoplasmaSchool of Studies in Microbiology
Ěý
This document summarizes a seminar presentation on Mycoplasma. Key points include:
- Mycoplasma are the smallest bacteria capable of autonomous growth. They lack cell walls and are sensitive to osmotic shock and antibiotics like tetracyclines.
- They were first discovered in 1843 and isolated from cattle pleural fluid. Techniques for culturing mycoplasma were developed in the late 19th century.
- Mycoplasma can take different shapes and sizes. They reproduce through binary fission or budding.
- They are classified as facultative anaerobes in the class Mollicutes. Some require sterols for growth while others do not. genetically modified organisms in the field of bio-remediation



genetically modified organisms in the field of bio-remediationswayam prakas nanda
Ěý
genetically modified organisms used for remediation of heavy metals, radioactive waste for bio-remediation Cot curve 



Cot curve EmaSushan
Ěý
Cot value and Cot Curve analysis is a technique for measuring DNA complexity based on renaturation kinetics. DNA is denatured and allowed to reanneal, with larger DNA taking longer. Cot value accounts for DNA concentration, time, and buffer effects, representing repetitive sequences - lower Cot means more repeats. Examples show bacteria have nearly all single-copy DNA, while mouse has varying proportions of single-copy, middle repetitive, and highly repetitive sequences. Cot curve analysis provides information on genome size, complexity, and proportions of sequence types.Similar to Microbial phylogeny (20)
Ap Chapter 26 Evolutionary History Of Biological Diversity



Ap Chapter 26 Evolutionary History Of Biological Diversitysmithbio
Ěý
- Phylogeny investigates the evolutionary history and relationships between species through analysis of fossil, molecular, and genetic data. Systematists use phylogenetic trees to depict these relationships.
- A phylogenetic tree represents hypotheses about evolutionary divergence. Each branch point indicates where two lineages diverged from a common ancestor. Tracing shared derived characteristics helps systematists determine which groupings form monophyletic clades.
- Molecular systematics uses comparisons of DNA and other molecules to infer relatedness and reconstruct evolutionary history over hundreds of millions of years, helping to extend phylogenies beyond what can be learned from fossils alone. Molecular clocks and neutral theory aim to date evolutionary events but have limitations.SYSTEMATICS.pdf



SYSTEMATICS.pdfNopiImrael
Ěý
1. The document discusses various methods used to classify living things, including morphology, developmental characteristics, evolutionary descent, anatomical resemblance, and molecular information such as DNA sequences.
2. Taxonomy is the scientific classification of organisms and systematics is the study of the evolutionary relationships between organisms.
3. DNA sequencing and comparing DNA strands between organisms has provided new insights into evolutionary relationships, with more similar DNA indicating closer relation.bacterial systematics in the diversity of bacteria



bacterial systematics in the diversity of bacteriatanvirastogi16
Ěý
This document provides an overview of bacterial systematics and taxonomy. It discusses the key concepts of classification, identification, nomenclature, and evolutionary relationships. The three main types of systematics are described as evolutionary, numerical, and phylogenetic. The importance of taxonomy is explained for effective communication, cataloging species, and aiding research. Key methods for bacterial classification discussed include phenotypic characterization, analysis of rRNA sequences, multilocus sequence analysis, restriction fragment length polymorphism, and fatty acid methyl ester profiling.Classification And Taxonomy Of The World



Classification And Taxonomy Of The WorldKim Moore
Ěý
Classification and taxonomy were established to create a system for grouping and naming organisms to make identification easier. Aristotle was the first to group organisms into plants and animals based on traits like movement. Carl Linnaeus further developed taxonomy by grouping organisms into a hierarchical system of kingdoms, which classified organisms based on increasingly specific characteristics. Taxonomy allows scientists to better identify, compare, and build knowledge about the millions of species in the world.2015 bioinformatics phylogenetics_wim_vancriekinge



2015 bioinformatics phylogenetics_wim_vancriekingeProf. Wim Van Criekinge
Ěý
This document discusses phylogenetic trees and phylogenetics. It begins with definitions of key terms like clade, species concepts, and branching order in phylogenetic trees. It provides examples of how phylogenetics is used in various fields like forensics, epidemiology, conservation biology, and pharmaceutical research. It also discusses choosing appropriate genetic sequences to use in phylogenetic analysis and highlights the pioneering work of Carl Woese in using rRNA sequences.2016 bioinformatics i_phylogenetics_wim_vancriekinge



2016 bioinformatics i_phylogenetics_wim_vancriekingeProf. Wim Van Criekinge
Ěý
This document provides an overview of phylogenetic methodologies. It defines key phylogenetic terms like clade, internal node, and outgroups. It discusses different species concepts and how phylogenetic trees illustrate evolutionary relationships. It also covers popular phylogenetic methodologies like distance methods, maximum parsimony, and maximum likelihood. Distance methods calculate pairwise distances and cluster sequences into trees. UPGMA averages these distances while neighbor joining finds the shortest branches. The document highlights the use of phylogenetic analysis across various fields.Classification



Classificationilanasaxe
Ěý
The document discusses biological classification and taxonomy. It introduces binomial nomenclature, which assigns organisms a two-part scientific name, as well as hierarchical classification systems. It also describes modern classification systems that incorporate data from fossils, physical traits, and DNA/RNA to construct phylogenetic trees representing evolutionary relationships among organisms.Bacterial taxonomy, pixel copy (2)

Bacterial taxonomy, pixel copy (2)Mwebaza Ivan
Ěý
This document discusses taxonomy and classification of organisms. It describes how taxonomy involves classifying organisms into groups based on similarities and evolutionary relationships. There are three domains of life - Archaea, Bacteria, and Eukarya. Bacteria are prokaryotic cells that are classified based on characteristics like cell structure, staining properties, shape, and biochemical reactions. Proper classification and naming of bacteria according to taxonomic rules is important for effective communication among microbiologists and organizing biological knowledge.Evolution and Phylogeny of fungi- An Evolutionary history of fungi



Evolution and Phylogeny of fungi- An Evolutionary history of fungisunilsuriya1
Ěý
The phylogeny of fungi is a fascinating and complex topic, reflecting the diversity and evolutionary history of this kingdom. Here are some key points:
Evolution and Phylogeny of Fungi:
Ancient Origins: Fungi are believed to have appeared around one billion years ago. The oldest known fungal fossils date back to the Silurian Period, about 440 million years ago1.
Monophyletic Group: Fungi are a monophyletic group, meaning all modern fungi can be traced back to a single common ancestor.
Molecular Phylogenetics: Advances in molecular phylogenetics, particularly since the 1990s, have greatly enhanced our understanding of fungal evolution. These studies often use the small subunit ribosomal RNA gene (SSU rRNA) and other protein-coding genes to construct phylogenetic trees1.
Phyla and Classes: The kingdom Fungi is divided into several phyla, including Ascomycota, Basidiomycota, Chytridiomycota, Zygomycota, and Glomeromycota, among others. Each phylum contains multiple classes and orders2.
Dikarya Subkingdom: The Ascomycota and Basidiomycota phyla form the subkingdom Dikarya, characterized by the presence of a dikaryotic stage in their life cycle.Lab 12 Building Phylogenies Objectives .docx



Lab 12 Building Phylogenies Objectives .docxDIPESH30
Ěý
Lab 12
Building Phylogenies
Objectives
In this laboratory exercise, you will examine six species of agaricomycetes and predict the evolutionary
relationships among them. After completing this exercise you will be able to
• define ancestral characteristics, derived characteristics, branch point, and phylogeny.
• predict ancestral and derived characteristics for agaricomycetes.
• construct a phylogeny (phylogenetic tree).
• support the phylogeny with data.
• explain how evolutionary biologists discover evolutionary relationships.
Introduction
One of the most compelling pieces of evidence for evolution is that organisms have amazing similarities. An
example that almost everyone has heard before is that the limbs of birds, bats, horses, moles, cats, frogs,
humans, turtles, and other vertebrates have virtually the same skeletal plan. Furthermore, even snakes and
whales show structural remnants of the limbs of their ancestors. The evolutionary interpretation of these
similarities is that the vertebrate limb has been modified by natural selection to perform different functions
(for example, running, digging, flying). Another commonly used example is that the embryos of turtles,
mice, humans, chickens, and many other vertebrates are amazingly similar. Furthermore, the proteins and
DNA of organisms are remarkably similar. Why, do you suppose, can human diabetics use insulin extracted
from pigs to control their blood sugar levels? Well, the reason is that the chemical structure of human and
pig insulin is very similar.
In addition to these similarities, we discover that organisms that appear similar in one respect are often
similar in other respects (we can say the patterns are “concordant”). For example, organisms that are
similar morphologically (in shape) have similar protein structures. Organisms that are less similar
morphologically have less similar protein structures. This pattern holds for traits that are not easily
modified by evolution, but not so often by traits that are easily modified by selection. For example, flower
color might not be a good trait to use when looking for concordance because it is easily changed
genetically.
The concordance of traits is an important support of evolution. Imagine that we saw that organisms similar
in one set of characteristics were very different in a second set of characteristics and different again in a
third set of characteristics. This situation would be chaotic and we would be forced to question the reality
1
of evolution. The development of methods of DNA and protein analysis has shown dramatically that
organisms that are similar morphologically are also similar at the genetic level.
So, similarity among organisms provides evidence for evolution. We can then turn around and use the
similarities to try to reconstruct evolutionary relationships. That is the purpose of today’s lab: to construct a
hypothes ...Phylogeny & classification



Phylogeny & classificationilanasaxe
Ěý
The document discusses the evolution of biological classification systems from Linnaeus' original system to modern phylogenetic classification based on evolutionary relationships. It introduces key concepts like binomial nomenclature, phylogeny, clades, homology, molecular clocks, and the three domain system of classifying life into Bacteria, Archaea, and Eukarya. Examples are provided to illustrate phylogenies and how to determine evolutionary relationships between organisms from phylogenetic trees.Bioinformatics t6-phylogenetics v2013-wim_vancriekinge



Bioinformatics t6-phylogenetics v2013-wim_vancriekingeProf. Wim Van Criekinge
Ěý
This document provides an overview of phylogenetics and the key concepts involved. It defines phylogenetics as reconstructing evolutionary relationships through phylogenetic trees. Species concepts and examples of using phylogenetics are discussed. The importance of the tree-of-life in illustrating how all life on Earth is related is explained. Important dates in the evolutionary history of life and commonly used gene sequences like rRNA for constructing phylogenetic trees are also summarized.Bioinformatica 24-11-2011-t6-phylogenetics



Bioinformatica 24-11-2011-t6-phylogeneticsProf. Wim Van Criekinge
Ěý
The document discusses the topic of phylogenetics. It begins with definitions of key terms like phylogeny, phylogenetic tree, clade, and orthologous genes. It then provides examples of how phylogenetic methods are used in fields like epidemiology, conservation biology, and pharmaceutical research. The document also discusses choosing appropriate genetic sequences to use in phylogenetic analysis and introduces molecular clock models.Bioinformatics t6-phylogenetics v2014



Bioinformatics t6-phylogenetics v2014Prof. Wim Van Criekinge
Ěý
This document provides an overview of phylogenetics and phylogenetic trees. It defines key terms like phylogeny, clade, species concepts, and the tree of life. It discusses using phylogenetic methods to solve crimes, study disease transmission, and aid conservation. Methods covered include distance methods, maximum likelihood, maximum parsimony, and software like PHYLIP. Ribosomal RNA genes are highlighted as useful sequences for constructing deep phylogenetic trees across all life.Biosystematics. 



Biosystematics. SabahatKhushnuma
Ěý
This document discusses biological systematics and taxonomy. It defines systematics as the scientific study of the diversity and classification of organisms, and the relationships between them. It notes that systematics involves classifying organisms into groups based on evolutionary relationships. The document also defines taxonomy as the scientific naming and classification of organisms according to a standardized system, and outlines the different levels of taxonomy from alpha to gamma. Microbial Phylogenomics (EVE161) Class 3: Woese and the Tree of Life



Microbial Phylogenomics (EVE161) Class 3: Woese and the Tree of LifeJonathan Eisen
Ěý
Microbial Phylogenomics (EVE161) at UC Davis Spring 2016. Co-taught by Jonathan Eisen and Holly Ganz.
Class 3: Era I: Woese and the Tree of Life Systematics vs taxonomy.pdf



Systematics vs taxonomy.pdfmohsinali52313
Ěý
Asslamoalaikum
here is an overview about systematics and its link with taxonomy.
contact for copyright.
KOUSIK_GHOSHPhenetics and Cladistics2020-04-05Phenetics and Cladistics.pptx



KOUSIK_GHOSHPhenetics and Cladistics2020-04-05Phenetics and Cladistics.pptxPriyankaChakraborty95
Ěý
Classification is the process of organizing taxa into groups based on their relationships. The main purposes of classification are for information storage and retrieval, to reflect evolutionary relationships, and to have a stable yet predictive system. There are different approaches to classification, including artificial, phenetic, cladistic, and numerical taxonomy methods. Phenetic classification groups taxa based on overall similarity regardless of evolutionary relationships, while cladistic classification aims to group taxa based on shared derived characteristics tracing back to a common ancestor.Recently uploaded (20)
ASP.NET Web API Interview Questions By Scholarhat



ASP.NET Web API Interview Questions By ScholarhatScholarhat
Ěý
ASP.NET Web API Interview Questions By ScholarhatInventory Reporting in Odoo 17 - Odoo 17 Inventory App



Inventory Reporting in Odoo 17 - Odoo 17 Inventory AppCeline George
Ěý
This slide will helps us to efficiently create detailed reports of different records defined in its modules, both analytical and quantitative, with Odoo 17 ERP.Functional Muscle Testing of Facial Muscles.pdf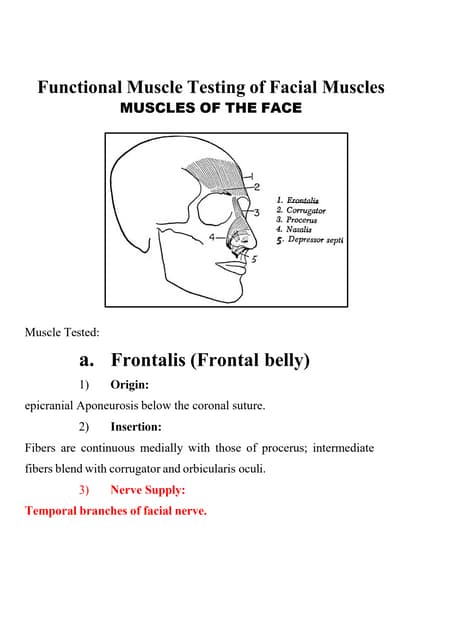



Functional Muscle Testing of Facial Muscles.pdfSamarHosni3
Ěý
Functional Muscle Testing of Facial Muscles.pdfFull-Stack .NET Developer Interview Questions PDF By ScholarHat



Full-Stack .NET Developer Interview Questions PDF By ScholarHatScholarhat
Ěý
Full-Stack .NET Developer Interview Questions PDF By ScholarHatComprehensive Guide to Antibiotics & Beta-Lactam Antibiotics.pptx



Comprehensive Guide to Antibiotics & Beta-Lactam Antibiotics.pptxSamruddhi Khonde
Ěý
📢 Comprehensive Guide to Antibiotics & Beta-Lactam Antibiotics
🔬 Antibiotics have revolutionized medicine, playing a crucial role in combating bacterial infections. Among them, Beta-Lactam antibiotics remain the most widely used class due to their effectiveness against Gram-positive and Gram-negative bacteria. This guide provides a detailed overview of their history, classification, chemical structures, mode of action, resistance mechanisms, SAR, and clinical applications.
📌 What You’ll Learn in This Presentation
âś… History & Evolution of Antibiotics
âś… Cell Wall Structure of Gram-Positive & Gram-Negative Bacteria
âś… Beta-Lactam Antibiotics: Classification & Subtypes
âś… Penicillins, Cephalosporins, Carbapenems & Monobactams
âś… Mode of Action (MOA) & Structure-Activity Relationship (SAR)
âś… Beta-Lactamase Inhibitors & Resistance Mechanisms
âś… Clinical Applications & Challenges.
🚀 Why You Should Check This Out?
Essential for pharmacy, medical & life sciences students.
Provides insights into antibiotic resistance & pharmaceutical trends.
Useful for healthcare professionals & researchers in drug discovery.
👉 Swipe through & explore the world of antibiotics today!
đź”” Like, Share & Follow for more in-depth pharma insights!Interim Guidelines for PMES-DM-17-2025-PPT.pptx



Interim Guidelines for PMES-DM-17-2025-PPT.pptxsirjeromemanansala
Ěý
This is the latest issuance on PMES as replacement of RPMS. Kindly message me to gain full access of the presentation. ASP.NET Interview Questions PDF By ScholarHat



ASP.NET Interview Questions PDF By ScholarHatScholarhat
Ěý
ASP.NET Interview Questions PDF By ScholarHatOral exam Kenneth Bech - What is the meaning of strategic fit?



Oral exam Kenneth Bech - What is the meaning of strategic fit?MIPLM
Ěý
Presentation of the CEIPI DU IPBA oral exam of Kenneth Bech - What is the meaning of strategic fit? Odoo 18 Accounting Access Rights - Odoo 18 şÝşÝߣs



Odoo 18 Accounting Access Rights - Odoo 18 şÝşÝߣsCeline George
Ěý
In this slide, we’ll discuss on accounting access rights in odoo 18. To ensure data security and maintain confidentiality, Odoo provides a robust access rights system that allows administrators to control who can access and modify accounting data. Azure Data Engineer Interview Questions By ScholarHat



Azure Data Engineer Interview Questions By ScholarHatScholarhat
Ěý
Azure Data Engineer Interview Questions By ScholarHatHow to Configure Proforma Invoice in Odoo 18 Sales



How to Configure Proforma Invoice in Odoo 18 SalesCeline George
Ěý
In this slide, we’ll discuss on how to configure proforma invoice in Odoo 18 Sales module. A proforma invoice is a preliminary invoice that serves as a commercial document issued by a seller to a buyer.How to create security group category in Odoo 17



How to create security group category in Odoo 17Celine George
Ěý
This slide will represent the creation of security group category in odoo 17. Security groups are essential for managing user access and permissions across different modules. Creating a security group category helps to organize related user groups and streamline permission settings within a specific module or functionality.Dr. Ansari Khurshid Ahmed- Factors affecting Validity of a Test.pptx



Dr. Ansari Khurshid Ahmed- Factors affecting Validity of a Test.pptxKhurshid Ahmed Ansari
Ěý
Validity is an important characteristic of a test. A test having low validity is of little use. Validity is the accuracy with which a test measures whatever it is supposed to measure. Validity can be low, moderate or high. There are many factors which affect the validity of a test. If these factors are controlled, then the validity of the test can be maintained to a high level. In the power point presentation, factors affecting validity are discussed with the help of concrete examples.BISNIS BERKAH BERANGKAT KE MEKKAH ISTIKMAL SYARIAH



BISNIS BERKAH BERANGKAT KE MEKKAH ISTIKMAL SYARIAHcoacharyasetiyaki
Ěý
BISNIS BERKAH BERANGKAT KE MEKKAH ISTIKMAL SYARIAHOne Click RFQ Cancellation in Odoo 18 - Odoo şÝşÝߣs



One Click RFQ Cancellation in Odoo 18 - Odoo şÝşÝߣsCeline George
Ěý
In this slide, we’ll discuss the one click RFQ Cancellation in odoo 18. One-Click RFQ Cancellation in Odoo 18 is a feature that allows users to quickly and easily cancel Request for Quotations (RFQs) with a single click.Microbial phylogeny
- 1. Microbial Phylogeny: Classification of species in superior taxa and construction of phylogenetic trees based on evolutionary relationships. It is bringing order to the diverse variety of organisms present in nature. So there are two general ways the classification can be constructed. First one is based on the morphological characters (phenetic classification) and second is based on evolutionary relationship (phylogenetic classification) Phenetic classification – It is based on the mutual similarity of their phenotypic characteristics. It does not provide any information about phylogenetic relations. Phylogenetic classification- These systems are based on evolutionary relationships rather than external appearance .The term phylogeny [Greek phylon, tribe or race, and genesis, generation or origin] refers to the evolutionary development of a species. It is based on the direct comparison of genetic materials and gene product.
- 2. Phylogenetic tree Phylogenetic relationships are illustrated in the form of branched diagrams or trees. A phylogenetic tree is a graph made of branches that connect nodes. The nodes represent taxonomic units such as species or genes; the external nodes, those at the end of the branches, represent living organisms. The tree may have a time scale, or the length of the branches may represent the number of molecular changes that have taken place between the two nodes. Finally, a tree may be unrooted or rooted. An unrooted tree simply represents phylogenetic relationships but does not provide an evolutionary path. Figure (a) shows that A is more closely related to C than it is to either B or D, but does not specify the common ancestor for the four species or the direction of change. In contrast, the rooted tree Figure (b) does give a node that serves as the common ancestor and shows the development of the four species from this root. Fig. 1 . Phylogenetic tree. a) unrooted tree, b) rooted tree.
- 3. Domains Advances in genomic DNA sequencing of the microorganisms, biologists are increasingly adapting the classification of living organisms that recognizes three domains, a taxonomic level higher than kingdom. Archaebacteria are in one domain, eubacteria in a second, and eukaryotes in the third. Domain Eukarya is subdivided into four kingdoms plants, animals, fungi, protists. Fig. 2 Three domains based on rRNA sequence analysis. Domain- Archaebacteria The term archaebacteria (Greek, archaio, ancient) refers to the ancient origin of this group of bacteria, which seem to have diverged very early from the eubacteria. They are inhabited mostly in extreme environments. The archaebacteria are grouped (based primarily on the environments in which they live) into three general categories methanogens, extremophiles and non extreme Archaebacteria.
- 4. Domain- Bacteria The Eubacteria are the most abundant organisms on earth. It plays critical roles like cycling carbon and sulfur. Much of the world's photosynthesis is carried out by eubacteria. However, certain groups of eubacteria are also responsible for many forms of disease. Domain- Eukarya It consists of four kingdoms. The first of which is protista, mostly unicellular organism like amoeba. The other three kingdoms are plants, fungi, animals. Multicellularity and sexuality are the two unique characters that differentiate from prokaryote and eukaryotes. Fig. 3 - Universal Phylogenetic Tree PHYLOGENIC RELATIONSHIP DETERMINING TECHNIQUE: A)Parsimony Analysis Phylogenetic relationships also can be estimated by techniques such as parsimony analysis. In this approach, relationships are determined by estimating the minimum number of sequence changes required to give the final sequences being compared. It is presumed that evolutionary change occurs along the shortest pathway with the fewest changes or steps from an ancestor to the organism in question.
- 5. B)Molecular chronometers This concept, first suggested by Zuckerkandl and Pauling (1965), which is based on thought that the sequences of many rRNAs and proteins gradually change over time without destroying or severely altering their functions. Changes increases with time linearly. If sequences of similar molecules from two organisms differs, it means that they diverged very long time ago. C)Oligonucleotide signature sequences The 16S rRNA of most major phylogenetic groups has one or more characteristic nucleotide sequences called oligonucleotide signatures. Oligonucleotide signature sequences are specific oligonucleotide sequences that occur in most or all members of a particular phylogenetic group. They are rarely or never present in other groups, even closely related ones. Thus signature sequences can be used to place microorganisms in the proper group. Polyphasic Taxonomy Studying phylogeny based on both genotypic and phenotypic information ranging from molecular characteristics to ecological characters.







