Paper - Muhammad Gulraj
- 1. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 1 Computational Bioinformatics & High Performance Computing Name: Muhammad Gulraj Muhammad.gulraj@yahoo.com Topic: Performance analysis of next generation DNA analysis using genome analysis toolkit - GATK
- 2. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 2 Abstract Deoxyribonucleic Acid (DNA) sequencing is the method of determining the precise order of ânucleotidesâ within a molecule. It includes any method or technology that is used to find the order of the four bases 1. adenine 2. guanine 3. cytosine 4. thymine In a strand of DNA. The arrival of rapid DNA sequencing procedures has greatly accelerated biological and medical research and discovery. GATK or Genome analysis tool kit is a software/tool used for the analysis sequencing data. There are many projects that are already going on in this field which give us the idea of genetic variations in human genes. These projects are however very limited in scope due to the computational infrastructure/power requirements for analysis of the DNA/genes. Genome analysis tool kit (GATK) is a very good tool which can be used in such scenarios for high- throughput data. The GATK enables us to use CPU and memory in an optimized manner and is very useful for distributed as well as parallelization. GATK is very robust and enables scientists to write efficient next generation sequencing tools e.g. â1000 genome projectâ. In recent times there are many platforms that have been developed for next generation sequencing â NGS but the problem with these platforms is that they focus on the analysis of a specific class/problem and lack flexibility. On the other hand Genome analysis tool kit â GATK uses Map-reduce by splitting data management structure from analysis and specific calculations.
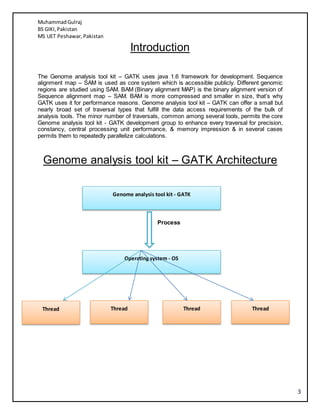
- 3. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 3 Introduction The Genome analysis tool kit â GATK uses java 1.6 framework for development. Sequence alignment map â SAM is used as core system which is accessible publicly. Different genomic regions are studied using SAM. BAM (Binary alignment MAP) is the binary alignment version of Sequence alignment map â SAM. BAM is more compressed and smaller in size, thatâs why GATK uses it for performance reasons. Genome analysis tool kit â GATK can offer a small but nearly broad set of traversal types that fulfill the data access requirements of the bulk of analysis tools. The minor number of traversals, common among several tools, permits the core Genome analysis tool kit - GATK development group to enhance every traversal for precision, constancy, central processing unit performance, & memory impression & in several cases permits them to repeatedly parallelize calculations. Genome analysis tool kit â GATK Architecture Process Genome analysis tool kit - GATK Operating system - OS Thread Thread Thread Thread
- 4. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 4 Description and Analysis The genome analysis tool kit â GATK distributes pool of mutual data arrangement schemes, known as traversals to developer/walkers for retrieving data for numerous analyses for example, calling Single Nucleotide Polymorphism â SNPs, counting the reads, building the base quality histograms, & recording the coverage of the sequencer reads over genome. Read based traversals offer a sequencer read and its related reference data during each repetition of the traversal. These traversals are distributed with reference base & associated reference ordered data. These repetitions are recurring correspondingly for every read or every reference base in the input Binary alignment MAP (BAM) file. Genome analysis tool kit â GATK (Infrastructure) Traversal Analysis Tool
- 5. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 5 The most puzzling features of NGS â (next generation sequencing) is handling the enormous scale of sequencing data. Logically shattering this prodigious group of information into controllable bits is very important for scalability, memory consumption, & operational parallelization of jobs. The GATK - genome analysis toolkit has engaged a new approach by distributing up data into multi kilo base parts, which can be called as shards. Shard sizes are calculated by the genome analysis toolkit â GATK. Shards are based on the fundamental storage features of the BAM - Binary alignment MAP & the system. Shards have the information for the related sub-region of the genome. Every shard is sub-divided by the traversal engine while feeding data to the walker. The genome analysis toolkit - GATK in these shards also manages reference ordered data with precise loci. The genome analysis toolkit - GATK is not restricted to the number of reference-ordered data tracks. Several analysis approaches are alarmed with calculation complete data for a single individual, collection, or group. Due to different reasons, data from next generation sequencing - NGS are not constantly arranged and clustered in the same style, which makes it difficult manages analyze a single data source and the probability of making an error is large. To resolve this issue, the genome analysis toolkit - GATK is proficient of merging various Binary alignments MAP - BAM files on the go, permitting numerous sequencing runs flawlessly into a solitary analysis without changing the input files. The compound sequencing data can be saved to disk, which makes this an effective means of merging data. The genome analysis toolkit - GATK offers several methods for the parallelization of jobs and issues. Using interval processing, programmers can divide jobs by genomic locations & give out every interval to a genome analysis toolkit - GATK case on a distributed system. The genome analysis toolkit - GATK provides automatic common memory parallelization, in which the GATK â genome analysis toolkit accomplishes several cases of the traversal engine & the walker. Walkers, which want to use this joint memory parallelization, implement the Tree Reducible interface due to which genome analysis toolkit can merge two reduce. The genome analysis toolkit also gathers the results from the individual walkers & combines. We have done the analyses using accessible data from the â1000-Genomes-Projectâ. This data is readily available from DCC - Data Coordination Center as BAM files. Finding the DoC or depth of coverage in a mix sequencing run is a computationally modest, but serious analysis tool. DoC computations have a significant role in precise CNV discovery & SNP. While computationally modest, the creation of this tool is usually snared with the provisions of sequence read based information. We have executed a DoC - depth of coverage walker in the Genome analysis toolkit to demonstrate the influence of the GATK, as well as to illustrate the ease of coding/programming to the GATK toolkit. The depth of coverage program has 83 lines of code.
- 6. MuhammadGulraj BS GIKI,Pakistan MS UET Peshawar,Pakistan 6 The walker accepts a list covering the base and emanates the size of the pileup. Users optionally eliminate reads of small mapping excellence, reads specified to be deletions at the current locus, and extra read cleaning criteria. Like all genome analysis toolkits based tools, the depth of coverage analysis can provide a list of regions to find coverage, adding the average coverage over every region. This competence is specifically valuable in quality control and calculation metrics for mix capture re-sequencing. This procedure can be used to enumerate sequencing consequences over composite or extremely variable regions. The lengthy MHC which is also known as major histocompatibility complex has problematic sequencer read alignments because of the increasing rate of genetic erraticism. Geno-typer calculates the posterior probability of every geno-type, assumed the pileup of sequencer reads that shield the current locus, and predictable hetero-zygosity of the sample. This calculation is used to originate the prior probability each of the likely 10 diploid genotypes. The algorithm was executed in the genome analysis toolkit, as a locus based walker. .We used the geno-typing algorithm to Pilot 3 deep coverage data for the CEU. On a solitary processor, this computation needs 867 minutes to process the 247,249,717 million loci of chromosome 1. Furthermore, the genome analysis toolkitâs built in provision for mutual memory parallelization permits us to rapidly add CPU (central processing unit) assets to minimize the run time of objective analyses. The passed time to geno-type drops almost exponentially through the accumulation of only 13 extra processing nodes, with no alteration to the code. This flexibility permits end users to allocate central processing unit on the importance of their completion, or to rapidly finish an analysis by allocating huge computing means to a single run. The genome analysis toolkit can be segregated to large computational clusters; this will help dropping elapsed time for end user analyses. This geno-typer achieves reasonably well at classifying variants with 83% in Single Nucleotide Polymorphism â SNP. The genome analysis toolkitâs efficiency and competence has allowed these tools to be effortlessly and quickly deployed as well processing 100âs of lanes in the production re-sequencing