Pcr
- 1. Dr. Hanumantappa B Nayaka M.Sc., M.Phil., PhD., PDF(Spain)., MIScT (U K) Asst. Professor Department of Life Sciences Kristu Jayanti College Autonomous Bengaluru-77, Karnataka PCR - Polymerase Chain Reaction
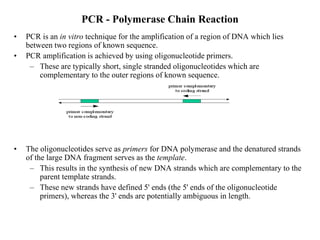
- 2. PCR - Polymerase Chain Reaction • PCR is an in vitro technique for the amplification of a region of DNA which lies between two regions of known sequence. • PCR amplification is achieved by using oligonucleotide primers. – These are typically short, single stranded oligonucleotides which are complementary to the outer regions of known sequence. • The oligonucleotides serve as primers for DNA polymerase and the denatured strands of the large DNA fragment serves as the template. – This results in the synthesis of new DNA strands which are complementary to the parent template strands. – These new strands have defined 5' ends (the 5' ends of the oligonucleotide primers), whereas the 3' ends are potentially ambiguous in length.
- 6. Primer selection • Primer is an oligonucleotide sequence – will target a specific sequence of opposite base pairing (A-T, G-C only) of single-stranded nucleic acids • For example, there is a – ¼ chance (4-1) of finding an A, G, C or T in any given DNA sequence; there is a – 1/16 chance (4-2) of finding any dinucleotide sequence (eg. AG); a – 1/256 chance of finding a given 4-base sequence. • Thus, a sixteen base sequence will statistically be present only once in every 416 bases (=4 294 967 296, or 4 billion): this is about the size of the human or maize genome, and 1000x greater than the genome size of E. coli.
- 7. Primer Specificity • Universal – amplifies ALL bacterial DNA for instance • Group Specific – amplify all denitrifiers for instance • Specific – amplify just a given sequence • Forward and reverse primers • If you know the sequence targeted for amplification, you know the size which the primers should be anealing across • If you don’t know the sequence… What do you get?
- 8. DNA Polymerase • DNA Polymerase is the enzyme responsible for copying the sequence starting at the primer from the single DNA strand • Commonly use Taq, an enzyme from the hyperthermophilic organisms Thermus aquaticus, isolated first at a thermal spring in Yellowstone National Park • This enzyme is heat-tolerant → useful both because it is thermally tolerant (survives the melting T of DNA denaturation) which also means the process is more specific, higher temps result in less mismatch – more specific replication
- 9. RFLP • Restriction Fragment Length Polymorphism • Cutting a DNA sequence using restriction enzymes into pieces → specific enzymes cut specific places Starting DNA sequence: 5’-TAATTTCCGTTAGTTCAAGCGTTAGGACC 3’-ATTAAAGGCAATCAAGTTCGCAATAATGG Enzyme X 5’-TTC- 3”-AAG- Enzyme X 5’-TTC- 3”-AAG- 5’-TAATTT 3’-ATTAAA 5’-CCGTTAGTT 3’-GGCAATCAA 5’-CAAGCGTTAGGACC 3’-GTTCGCAATAATGG
- 10. RFLP • DNA can be processed by RFLP either directly (if you can get enough DNA from an environment) or from PCR product • T-RFLP (terminal-RFLP) is in most respects identical except for a marker on the end of the enzyme • Works as fingerprinting technique because different organisms with different DNA sequences will have different lengths of DNA between identical units targeted by the restriction enzymes – specificity can again be manipulated with PCR primers
- 11. Electrophoresis • Fragmentation products of differing length are separated – often on an agarose gel bed by electrophoresis, or using a capilarry electrophoretic separation