Polymerase Chain Reaction
- 2. INTRODUCTION ? PCR, polymerase chain reaction, is an in-vitro technique for amplification of a region of DNA whose sequence is known or which lies between two regions of known sequence ? PCR can be regarded as a form of molecular cloning since it is a technique similar to DNA replication that occurs in the cells
- 3. REACTION COMPONENTS ? DNA template ? Primers ? Enzyme ? dNTPs ? Buffers
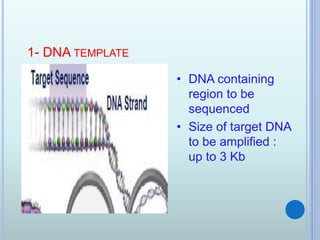
- 4. 1- DNA TEMPLATE ? DNA containing region to be sequenced ? Size of target DNA to be amplified : up to 3 Kb
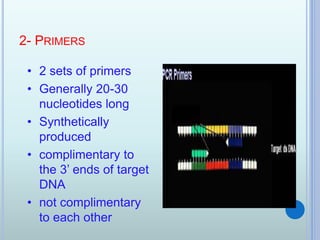
- 5. 2- PRIMERS ? 2 sets of primers ? Generally 20-30 nucleotides long ? Synthetically produced ? complimentary to the 3ĄŊ ends of target DNA ? not complimentary to each other
- 6. 3-ENZYME ? Usually Taq Polymerase or anyone of the natural or Recombinant thermostable polymerases ? Stable at T0 up to 950 C ? High processivity
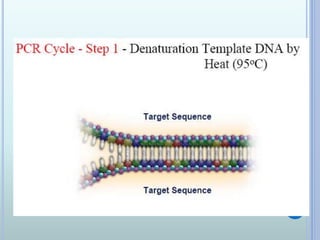
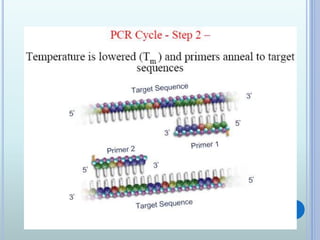
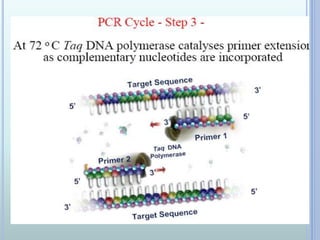
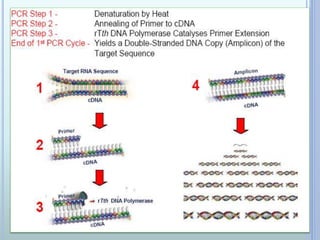
- 7. THE PCR CYCLE Contains of 3 steps: - Denaturation of DNA at 950C - Primer hybridization ( annealing) at 40- 500C - DNA synthesis ( Primer extension) at 720C
- 12. TARGET AMPLIFICATION ? The target DNA is amplified into multiple copies in a exponential pattern with repeated cycles of PCR and multiple copies of the DNA are produced
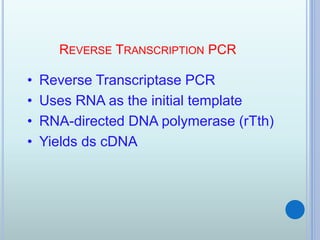
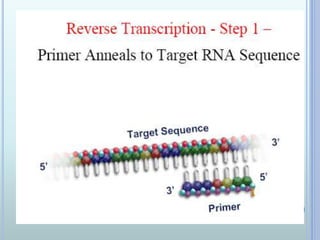
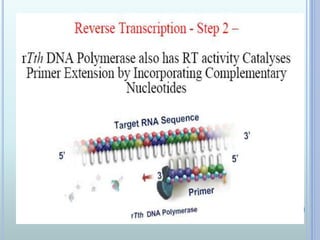
- 13. REVERSE TRANSCRIPTION PCR ? Reverse Transcriptase PCR ? Uses RNA as the initial template ? RNA-directed DNA polymerase (rTth) ? Yields ds cDNA
- 17. DETECTION OF AMPLIFICATION PRODUCTS ? Gel electrophoresis ? Sequencing of amplified fragment ? Southern blot ? etc...
- 18. APPLICATIONS ? Amplification of small amounts of DNA for further analysis by DNA fingerprinting. ? The analysis of ancient DNA from fossils. ? Mapping the human (and other species) genome. ? The isolation of a particular gene of interest from a tissue sample. ? Generation of probes: large amount of probes can be synthesized by this technique. ? Production of DNA for sequencing: Target DNA in clone is amplified using appropriate primers and then its sequence determined. Helpful in conditions where amount of DNA is small.
- 19. ? Analysis of mutations: Deletions and insertions in a gene can be detected by differences in size of amplified product. ? Diagnosis of monogenic diseases (single gene disorders): For pre-natal diagnosis, PCR is used to amplify DNA from foetal cells obtained from amniotic fluid. PCR has also proved very important in carrier testing. ? Detection of microorganisms: Especially of organisms and viruses that are difficult to culture or take long time to culture or dangerous to culture.
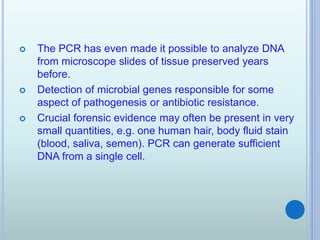
- 20. ? The PCR has even made it possible to analyze DNA from microscope slides of tissue preserved years before. ? Detection of microbial genes responsible for some aspect of pathogenesis or antibiotic resistance. ? Crucial forensic evidence may often be present in very small quantities, e.g. one human hair, body fluid stain (blood, saliva, semen). PCR can generate sufficient DNA from a single cell.
- 21. ADVANTAGES ? Automated, fast, reliable (reproducible) results ? Contained :(less chances of contamination) ? High output ? Sensitive ? Broad uses ? Defined, easy to follow protocols
- 22. ? Real-time polymerase chain reaction, also called quantitative real time polymerase chain reaction is a laboratory technique based on the PCR, which is used to amplify and simultaneously quantify a targeted DNA molecule. Real-time PCR
- 23. ? Real-Time PCR permits the analysis of the products while the reaction is actually in progress. This is achieved by using various fluorescent dyes which react with the amplified product and can be measured by an instrument. This also facilitates the quantitation of the DNA. ? Quantitative PCR (Q-PCR), as this technique is known, is used to measure the quantity of a PCR product
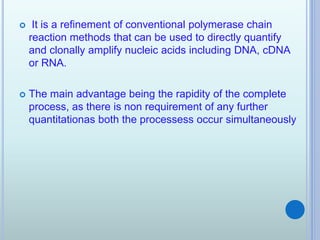
- 24. ? It is a refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids including DNA, cDNA or RNA. ? The main advantage being the rapidity of the complete process, as there is non requirement of any further quantitationas both the processess occur simultaneously
- 25. PROCESS OF QUANTIFICATION OF DNA ? A DNA binding dye such as SYBR green is introduced into the reaction mixture of PCR. ? Then the PCR cycles are started. ? As amplicons accumulate, SYBR green binds to the ds DNA proportionally. ? Fluorescence emission of the dye is detected following excitation.
- 26. ? The binding SYBR green is non specific. Therefore, in order to detect specific amplicons an oligonucleotide probe labeled with a fluorescent reporter and quencher molecule at either end is included in the reaction in the place of SYBR green. ? When the oligonucleotide probe binds to the target sequence, the 5ĄŊ exonuclease activity of Taq polymerase degrades and releases the reporter from the quencher.
- 27. ? A signal is generated which increases in direct proportion in no. of starting molecule. ? Hence the detection system is able to induce and detect fluorescence in real time as the PCR proceeds.
- 28. APPLICATIONS ? Gene expression analysis ? Cancer research ? Drug research ? Disease diagnosis and management ? Viral quantification ? Food testing ? Percent GMO food ? Animal and plant breeding ? Gene copy number
- 29. REFERENCES ? Molecular Biology and Biotechnology by, Walker and Rapley, 5th edition, pg no. ĻC 122- 128 ? Pharmaceutical Biotechnology by , Chandrakant Kokare, Pg no. -10.15 ? A textbook of biotechnology by R. C. Dubey ? International Journal of Biomedical Research, review article Ą°POLYMERASE CHAIN REACTION: METHODS, PRINCIPLES AND APPLICATIONĄą by, Dr. Mohini Joshi & Dr. Deshpande. J. D
- 30. THANK YOU