EM 3D reconstruction
- 1. Cryo-EM 3D reconstruction Fan Zhitao NUS Graduate School February 20, 2014
- 2. Images are everywhere In our life In research Z.T. Fan °™ Cryo-EM 3D reconstruction 2/11
- 3. EM imaging process Z.T. Fan °™ Cryo-EM 3D reconstruction 3/11
- 4. The EM images Z.T. Fan °™ Cryo-EM 3D reconstruction 4/11
- 5. Mathematical modeling The mathematical model is Af = g Z.T. Fan °™ Cryo-EM 3D reconstruction 5/11
- 6. Challenges Challenges: large noise large data: 100, 000 projections with size 512 A is hard to write out Contribution Proposed a memory-saving tight wavelet frame based algorithm Done the convergence analysis of this algorithm Z.T. Fan °™ Cryo-EM 3D reconstruction 6/11
- 7. Sparse representation The image has a sparse representation under wavelet system. If we have a sparse representation, we may formulate a mathematical model: min g ? Af f 2 + ¶À Wf 1 W is the discrete wavelet transform. Dong and Shen 2005, Ron and Shen 1995 Z.T. Fan °™ Cryo-EM 3D reconstruction 7/11
- 8. The algorithm Algorithm 1 fk+1 = (I ? ?A A)W T¶À Wfk + ?A g The advantage Simple: one soft-thresholding and one gradient descent Small memory footprint: one wavelet transform Z.T. Fan °™ Cryo-EM 3D reconstruction 8/11
- 9. Simulated data: E. coli ribosome Simulated 2D noisy projections 3D reconstruction Ground truth Z.T. Fan °™ Cryo-EM 3D reconstruction BP Proposed algorithm Experiment results 9/11
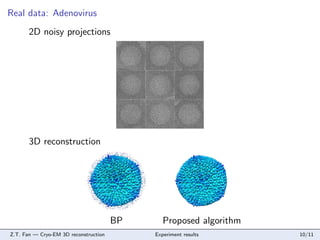
- 10. Real data: Adenovirus 2D noisy projections 3D reconstruction BP Z.T. Fan °™ Cryo-EM 3D reconstruction Proposed algorithm Experiment results 10/11
- 11. Thank you! Special thanks to DR Li Ming, Chinese Academy of Science Prof Ji Hui, NUS mathematics Prof Shen Zuowei, NUS mathematics Z.T. Fan °™ Cryo-EM 3D reconstruction Acknowledgement 11/11