Marco Andrello - Incongruency between model-based and genetic-based estimates of effective population size
- 1. Incongruency between demographic-based and genetic-based estimates of effective population size Marco Andrello , M Gaudeul, I Till-Bottraud and O.E Gaggiotti Mod├©les en Ecologie Evolutive, Montpellier, 31st May - 2nd June 2010
- 2. Definition Use of N e in conservation In evolutionary ecology Estimation methods for N e Assessment of estimators: different approaches give inconsistent estimates Why??
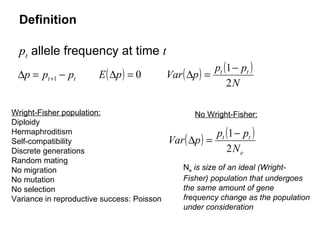
- 3. Definition p t allele frequency at time t Wright-Fisher population: Diploidy Hermaphroditism Self-compatibility Discrete generations Random mating No migration No mutation No selection Variance in reproductive success: Poisson N e is size of an ideal (Wright-Fisher) population that undergoes the same amount of gene frequency change as the population under consideration No Wright-Fisher:
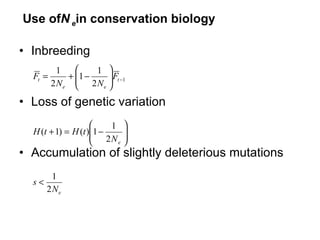
- 4. Use of N e in conservation biology Inbreeding Loss of genetic variation Accumulation of slightly deleterious mutations
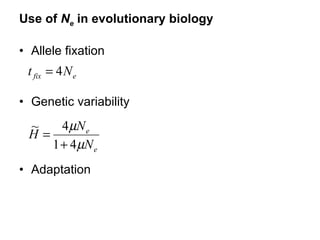
- 5. Use of N e in evolutionary biology Allele fixation Genetic variability Adaptation
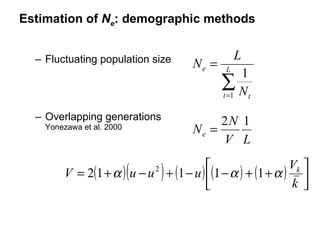
- 6. Estimation of N e : demographic methods Fluctuating population size Overlapping generations Yonezawa et al. 2000
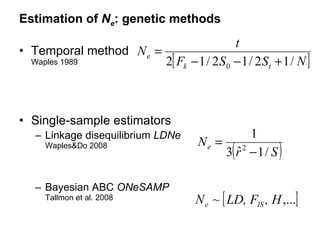
- 7. Estimation of N e : genetic methods Temporal method Waples 1989 Single-sample estimators Linkage disequilibrium LDNe Waples&Do 2008 Bayesian ABC ONeSAMP Tallmon et al. 2008
- 8. Do the two approaches give the same estimates? Eryngium alpinum Overlapping generations Stage structure Variance in reproductive success Demographic and genetic data for 4 populations: 2001 ŌĆō 2009 demographic survey Genetic samples; 8 microsatellites Ne estimation: LDNe ONeSAMP Demographic Yonezawa et al. 2000
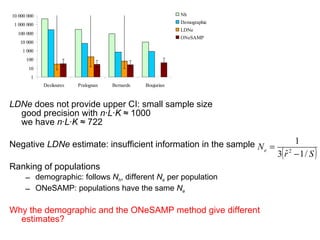
- 9. LDNe does not provide upper CI: small sample size good precision with n┬ĘL┬ĘK Ōēł 1000 we have n┬ĘL┬ĘK Ōēł 722 Negative LDNe estimate: insufficient information in the sample Ranking of populations demographic: follows N h , different N e per population ONeSAMP: populations have the same N e Why the demographic and the ONeSAMP method give different estimates?
- 10. Demographic and genetic methods might measure different things Potential biases of the demographic method Bias of genetic methods?
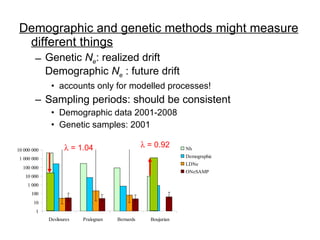
- 11. Demographic and genetic methods might measure different things Genetic N e : realized drift Demographic N e : future drift accounts only for modelled processes! Sampling periods: should be consistent Demographic data 2001-2008 Genetic samples: 2001 ’ü¼ = 1.04 ’ü¼ = 0.92
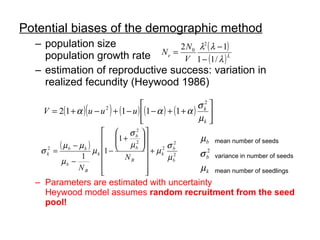
- 12. Potential biases of the demographic method population size population growth rate estimation of reproductive success: variation in realized fecundity (Heywood 1986) Parameters are estimated with uncertainty Heywood model assumes random recruitment from the seed pool! mean number of seeds variance in number of seeds mean number of seedlings
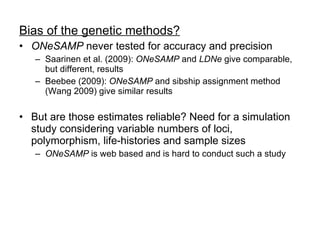
- 13. Bias of the genetic methods? ONeSAMP never tested for accuracy and precision Saarinen et al. (2009): ONeSAMP and LDNe give comparable, but different, results Beebee (2009): ONeSAMP and sibship assignment method (Wang 2009) give similar results But are those estimates reliable? Need for a simulation study considering variable numbers of loci, polymorphism, life-histories and sample sizes ONeSAMP is web based and is hard to conduct such a study
- 14. Conclusions and perspectives N e is an important parameter, but still difficult to estimate Demographic and genetic methods gave inconsistent results Use large sample size to get precise genetic estimates Conduct an assessment of ONeSAMP Extend comparisons to more species for which demographic data are available Use different methods when possible!