RDA WG proposal on connecting data policies, standards & databases in life sciences
Download as PPTX, PDF1 like1,466 views
RDA Plenary 3 (Dublin) WG proposal from Susanna Sansone (Biosharing), Simon Hodson (CO-DATA) and Rebecca Lawrence (F1000Research) on connecting data standards, policies and repositories in the life sciences.
1 of 8
Download to read offline








Recommended
The CATE Project



The CATE ProjectKehan Harman
Ěý
Presentation by Simon Mayo at the KikForum
Abstract:
As part of the CATE project we are developing keys in Lucid 3 to the genera of Araceae, and to the genus Anthurium (ca. 800spp.), Arum and Philodendron. The key to Arum is already online. These keys will be incorporated into a web-based taxonomic revision of the Araceae family as the plant model group for the project. Anthurium presents a particular challenge as it is a very large and difficult genus, within which it is currently nearly impossible for non-specialists to determine plants to species. We hope the key will go some way to solving this problem.BioSharing WG - ELIXIR IG - RDA Plenary 7, Tokyo, March 2016



BioSharing WG - ELIXIR IG - RDA Plenary 7, Tokyo, March 2016Susanna-Assunta Sansone
Ěý
Update on the BioSharing WG activities at the joint ELIXIR IG and BioSharing WG breakout: https://rd-alliance.org/joint-meeting-ig-elixir-bridging-force-wg-biosharing-registry.htmlPelajar cemerlang assignment computer app



Pelajar cemerlang assignment computer appFatin Fatihah
Ěý
Dokumen memberikan 7 teknik untuk mempertahankan citra keberhasilan pelajar, termasuk mengingat pencapaian masa lalu, berteman dengan orang berfikiran positif, menjauhi orang negatif, memulai dengan hal-hal kecil, membina hubungan baik dengan guru, mengurangi tekanan perasaan, dan berdoa.Morningcall



MorningcallGloblex Securities
Ěý
The document provides a market summary and commentary for October 20, 2011. It summarizes that the SET closed down 1.53% at 938.19 points on lower volume, with technical support seen at 923/914 and resistance at 973/990. Most world indices also declined except for the FTSE and DAX. Commodity prices fell while the baht strengthened against the dollar. The report also comments on various Thai companies and economic indicators.RDA Plenary6 bio_sharing_leaflet



RDA Plenary6 bio_sharing_leafletPeter McQuilton
Ěý
A 2-page leaflet detailing the life science database, standard, and policy registries in BioSharing, and the ability to make a Collection of these resources.BioSharing - mapping the landscape of Standards, Databases and Data policies ...



BioSharing - mapping the landscape of Standards, Databases and Data policies ...Peter McQuilton
Ěý
A 35 minute lecture presented at NETTAB2016 in Rome, Italy, 26th October, under the auspices of COST-CHARME and EMBNET.RDA Publishing Workflows



RDA Publishing WorkflowsPeter McQuilton
Ěý
This document discusses the BioSharing registry, which connects standards, databases, and policies in the life sciences. BioSharing provides a searchable portal for standards and databases, helping researchers choose the right options for publishing and funding requirements. It monitors the development of standards and their adoption. The registry links three sections on standards, databases, and policies to help answer common questions about which options to use. Users can search, filter, and refine results or create customized collections. BioSharing aims to support better informed decisions across the life sciences research community.Cross-linked metadata standards, repositories and the data policies - The Bio...



Cross-linked metadata standards, repositories and the data policies - The Bio...Peter McQuilton
Ěý
A 20 minute presentation given in Denver (CO) on the 17th September as part of the Biosharing Registry WG, Metadata Standards Catalog WG, and Publishing Data Workflows WG joint session at the Research Data Alliance 8th Plenary (part of International Data Week).
This presentation covers the explosion of metadata standards and databases in the life, biomedical and environmental sciences and how BioSharing is helping to understand this landscape, both in terms of the relationship between standards and other standards and databases, and the life cycle and evolution of each resource. BioSharing also links these resources to the data policies that recommend them (for example, from funding agencies or journal publishers), enabling an understanding of the entire data cycle, from conception to publishing and storage.RDA BioSharing WG + RDA Metabolomics IG OVERVIEWS



RDA BioSharing WG + RDA Metabolomics IG OVERVIEWSSusanna-Assunta Sansone
Ěý
Brief overviews of RDA BioSharing WG and RDA Metabolomics IG for RDA Pleanary 4; IGs and WGs chair session (Sunday) and ELIXIR IG breakout (Tuesday)Using community-defined metadata standards in the FAIR principles: how BioSha...



Using community-defined metadata standards in the FAIR principles: how BioSha...Peter McQuilton
Ěý
A 10 minute presentation given in Denver (CO) on the 16th September as part of the IG Elixir Bridging Force and Biosharing Registry WG joint session at the Research Data Alliance 8th Plenary (part of International Data Week).
This presentation covers the use of community-defined metadata standards in the life science, making these standards FAIR, and how BioSharing can help.RDA Webinar - BioSharing - mapping the landscape of data standards, repositor...



RDA Webinar - BioSharing - mapping the landscape of data standards, repositor...Peter McQuilton
Ěý
A 30 minute webinar presented on behalf of the RDA/Force11 BioSharing WG, covering our work to map data standards, databases, and data policies in the life, biomedical and environmental sciences.BioSharing şÝşÝߣs - Repository Fringe Edinburgh August 2015



BioSharing şÝşÝߣs - Repository Fringe Edinburgh August 2015Peter McQuilton
Ěý
The document describes a web-based portal that registers, links, and allows discovery of biological standards and databases. It monitors the development and evolution of standards and their use in databases and adoption in data policies. Its mission is to provide support and guidance to researchers, developers, curators, journal publishers, and funders on choosing the appropriate standards and databases by registering, linking, and making discoverable relevant biological standards and databases.ELIXIR Webinar: BioSharing



ELIXIR Webinar: BioSharingPeter McQuilton
Ěý
This document provides an overview of BioSharing.org, a portal that monitors and curates standards, databases, and data policies to help inform and educate users. It summarizes key features including tracking over 225 content standards, 115 databases, and 554 data policies. The portal aims to help users understand how standards are used and make informed decisions on standards selection. It also links standards to training materials and allows custom collections and recommendations to be created.FAIRsharing and DataCite: Data Repository Selection- Criteria That Matter



FAIRsharing and DataCite: Data Repository Selection- Criteria That MatterSusanna-Assunta Sansone
Ěý
Through a collaboration with Datacite, FAIRsharing is working with a number of journal publishers (PLOS, Springer Nature, F1000, Wiley, Taylor and Francis, Elsevier, EMBO Press, eLife, GigaScience and Cambridge University Press) to identify a common set of criteria for selecting and recommending data repositories (and associated standards) that will be implemented in FAIRsharing. Details of this work and participants at https://osf.io/m2bceOSFair2017 Workshop | FAIRSharing



OSFair2017 Workshop | FAIRSharingOpen Science Fair
Ěý
Rafael Jimenez presents the FAIRSharing on behalf of Peter McQuilton, Susanna-Assunta Sansone & the FAIRsharing team | OSFair2017 Workshop
Workshop title: How FAIR friendly is your data catalogue?
Workshop overview:
This workshop will build upon the work planned by the EOSCpilot data interoperability task and the BlueBridge workshop held on April 3 at the RDA meeting. We will investigate common mechanisms for interoperation of data catalogues that preserve established community standards, norms and resources, while simplifying the process of being/becoming FAIR. Can we have a simple interoperability architecture based on a common set of metadata types? What are the minimum metadata requirements to expose FAIR data to EOSC services and EOSC users?
DAY 3 - PARALLEL SESSION 6 & 7BioSharing - RDA Plenary 6 - Metadata Standards Catalog WG and BioSharing WG ...



BioSharing - RDA Plenary 6 - Metadata Standards Catalog WG and BioSharing WG ...Peter McQuilton
Ěý
An introduction to the metadata landscape in the life sciences. Covering metadata standards, the databases that implement them and the policies that endorse/recommend both standards and databases in the life sciences."Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013



"Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013Susanna-Assunta Sansone
Ěý
Overview of the landscape of standards in life sciences for the NIH BD2K
"Frameworks for Community-Based Standards Efforts" workshop
September 25, 2013 - September 26, 2013
Co-Chairs: Susanna Sansone, PhD and David Kennedy PhD.
The overall goal of this workshop is to learn what has worked and what has not worked in community-based standards efforts. Participants will have experience in leading specific community based standards initiatives. Prior to the workshop, participants will be asked to address in writing answers to specific questions regarding formulating, conducting, and maintaining such efforts. This information will be used to facilitate focused and actionable discussion at the workshop. Issuance of a Request for Information soliciting comment from the broader community on some of the key issues addressed in the workshop is currently envisioned.
Contact: BD2Kworkshops@mail.nih.gov
Agenda: Frameworks for Community-Based Standards Efforts (PDF 40.7KB)
Participant List: Roster of Invited Participants (PDF 32KB)
Forum (Join the discussion): http://frameworks.prophpbb.com
Watch Live: http://videocast.nih.gov/summary.asp?live=13088 - See more at: http://bd2k.nih.gov/workshops.html#cbseOverview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014



Overview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014Susanna-Assunta Sansone
Ěý
What to know when planning for your data management strategy and preparing a data management statement for a research proposal for BBSRC DTP first year studentsThe BioSharing portal - linking journal and funder data policies to databases...



The BioSharing portal - linking journal and funder data policies to databases...Peter McQuilton
Ěý
A 20 minute talk on the BioSharing portal, focusing on our work to link journal and funder data policies to the databases and data standards that they recommend/endorse. This was presented as part of a session on data policies in the life sciences with representation from JISC and Springer Nature.FAIR data and NPG Scientific Data: RIKEN Yokohama, 25 June, 2014



FAIR data and NPG Scientific Data: RIKEN Yokohama, 25 June, 2014Susanna-Assunta Sansone
Ěý
The document summarizes a presentation about making scientific data FAIR (Findable, Accessible, Interoperable, Reusable). It discusses the concept of FAIR data and several of the presenter's related projects. Examples are provided of using standards like ISA-Tab to structure metadata and make datasets interoperable. The presentation outlines the presenter's roles in data capture, publication, and standards development efforts to promote FAIR data principles. Scientific Data, a new journal for peer-reviewed data descriptions, is introduced as a way to make datasets more discoverable and reusable.BioSharing - Mapping the landscape of Standards, Database and Data Policies i...



BioSharing - Mapping the landscape of Standards, Database and Data Policies i...Peter McQuilton
Ěý
A 20 minute talk on BioSharing, presented at the COST CHARME data standards conference in Warsaw, Poland, on June 21th 2016NIH iDASH meeting on data sharing - BioSharing, ISA and Scientific Data



NIH iDASH meeting on data sharing - BioSharing, ISA and Scientific DataSusanna-Assunta Sansone
Ěý
1) The document discusses Susanna-Assunta Sansone's roles and work related to promoting FAIR data standards and practices.
2) It highlights some of her leadership positions with organizations like BioSharing that work to map and promote standards.
3) The document also discusses Scientific Data, a peer-reviewed journal launched by Nature Publishing Group to publish detailed descriptions of scientifically valuable datasets to facilitate reuse.BioSharing, an ELIXIR Interoperability Platform resource



BioSharing, an ELIXIR Interoperability Platform resourcePeter McQuilton
Ěý
A 20 minute presentation given at the 9th RDA Plenary in Barcelona as part of the BioSharing WG - ELIXIR Bridging Force IG session. This presentation covers the basics of what BioSharing is, who it's for, and how it captures and connects information on data standards, databases and data policies from the life, biomedical and environmental sciences.The BioSharing portal - linking databases, data standards and policies in the...



The BioSharing portal - linking databases, data standards and policies in the...Peter McQuilton
Ěý
A 15 minute presentation for the Interest Group on Agricultural Data (IGAD) RDA pre-meeting meeting. Presented in Barcelona (ES) on Monday 3rd April, 2017.Managing Big Data - Berlin, July 9-10, 201.



Managing Big Data - Berlin, July 9-10, 201.Susanna-Assunta Sansone
Ěý
Susanna-Assunta Sansone is a data consultant and honorary academic editor who works on several projects related to making data FAIR (Findable, Accessible, Interoperable, Reusable). She is the associate director of Scientific Data, a peer-reviewed journal focused on publishing data descriptors to describe and provide access to scientifically valuable datasets. The goal of Scientific Data is to help promote open science and data reuse by publishing structured metadata and narratives about datasets alongside traditional research articles.Brief History of Public Health Systems and Services Research (PHSSR) 



Brief History of Public Health Systems and Services Research (PHSSR) National Coordinating Center for Public Health Services & Systems Research
Ěý
The document provides a history of Public Health Services and Systems Research (PHSSR) and the National Coordinating Center. It discusses how early work included developing data standards, funding research through mini-grants, and convening networking opportunities. It also summarizes how the coordinating center aims to advance the field through activities like establishing an endnote library, supporting practice-based research networks, and increasing communication and visibility of PHSSR through various channels. In closing, it reflects on lessons learned about the challenges of cross-sector coordination and promoting underrepresented areas of research like PHSSR.Large Language Models (LLMs) part one.pptx



Large Language Models (LLMs) part one.pptxharmardir
Ěý
**The Rise and Impact of Large Language Models (LLMs)**
**Introduction**
In the rapidly evolving landscape of artificial intelligence (AI), one of the most groundbreaking advancements has been the development of Large Language Models (LLMs). These AI systems, trained on massive amounts of text data, have demonstrated remarkable capabilities in understanding, generating, and manipulating human language. LLMs have transformed industries, reshaped the way people interact with technology, and raised ethical concerns regarding their usage. This essay delves into the history, development, applications, challenges, and future of LLMs, providing a comprehensive understanding of their significance.
**Historical Background and Development**
The foundation of LLMs is built on decades of research in natural language processing (NLP) and machine learning (ML). Early language models were relatively simple and rule-based, relying on statistical methods to predict word sequences. However, the emergence of deep learning, particularly the introduction of neural networks, revolutionized NLP. The introduction of recurrent neural networks (RNNs) and long short-term memory (LSTM) networks in the late 1990s and early 2000s allowed for better sequential data processing.
The breakthrough moment for LLMs came with the development of Transformer architectures, introduced in the seminal 2017 paper "Attention Is All You Need" by Vaswani et al. The Transformer model enabled more efficient parallel processing and improved context understanding. This led to the creation of models like BERT (Bidirectional Encoder Representations from Transformers) and OpenAI’s GPT (Generative Pre-trained Transformer) series, which have since set new benchmarks in AI-driven text generation and comprehension.
**Core Mechanisms of LLMs**
LLMs rely on deep neural networks trained on extensive datasets comprising books, articles, websites, and other textual resources. The training process involves:
1. **Tokenization:** Breaking down text into smaller units (words, subwords, or characters) to be processed by the model.
2. **Pretraining:** The model learns general language patterns through unsupervised learning, predicting missing words or the next sequence in a text.
3. **Fine-tuning:** Adjusting the model for specific tasks, such as summarization, translation, or question-answering, using supervised learning.
4. **Inference:** The trained model generates text based on user input, leveraging probabilistic predictions to produce coherent responses.
Through these mechanisms, LLMs can perform a wide range of linguistic tasks with human-like proficiency.
**Applications of LLMs**
LLMs have found applications across various domains, including but not limited to:
1. **Content Generation:** LLMs assist in writing articles, blogs, poetry, and even code, helping content creators enhance productivity.
2. **Customer Support:** Chatbots and virtual assistants powered by LLMs provide automated yet contMore Related Content
Similar to RDA WG proposal on connecting data policies, standards & databases in life sciences (20)
RDA Publishing Workflows



RDA Publishing WorkflowsPeter McQuilton
Ěý
This document discusses the BioSharing registry, which connects standards, databases, and policies in the life sciences. BioSharing provides a searchable portal for standards and databases, helping researchers choose the right options for publishing and funding requirements. It monitors the development of standards and their adoption. The registry links three sections on standards, databases, and policies to help answer common questions about which options to use. Users can search, filter, and refine results or create customized collections. BioSharing aims to support better informed decisions across the life sciences research community.Cross-linked metadata standards, repositories and the data policies - The Bio...



Cross-linked metadata standards, repositories and the data policies - The Bio...Peter McQuilton
Ěý
A 20 minute presentation given in Denver (CO) on the 17th September as part of the Biosharing Registry WG, Metadata Standards Catalog WG, and Publishing Data Workflows WG joint session at the Research Data Alliance 8th Plenary (part of International Data Week).
This presentation covers the explosion of metadata standards and databases in the life, biomedical and environmental sciences and how BioSharing is helping to understand this landscape, both in terms of the relationship between standards and other standards and databases, and the life cycle and evolution of each resource. BioSharing also links these resources to the data policies that recommend them (for example, from funding agencies or journal publishers), enabling an understanding of the entire data cycle, from conception to publishing and storage.RDA BioSharing WG + RDA Metabolomics IG OVERVIEWS



RDA BioSharing WG + RDA Metabolomics IG OVERVIEWSSusanna-Assunta Sansone
Ěý
Brief overviews of RDA BioSharing WG and RDA Metabolomics IG for RDA Pleanary 4; IGs and WGs chair session (Sunday) and ELIXIR IG breakout (Tuesday)Using community-defined metadata standards in the FAIR principles: how BioSha...



Using community-defined metadata standards in the FAIR principles: how BioSha...Peter McQuilton
Ěý
A 10 minute presentation given in Denver (CO) on the 16th September as part of the IG Elixir Bridging Force and Biosharing Registry WG joint session at the Research Data Alliance 8th Plenary (part of International Data Week).
This presentation covers the use of community-defined metadata standards in the life science, making these standards FAIR, and how BioSharing can help.RDA Webinar - BioSharing - mapping the landscape of data standards, repositor...



RDA Webinar - BioSharing - mapping the landscape of data standards, repositor...Peter McQuilton
Ěý
A 30 minute webinar presented on behalf of the RDA/Force11 BioSharing WG, covering our work to map data standards, databases, and data policies in the life, biomedical and environmental sciences.BioSharing şÝşÝߣs - Repository Fringe Edinburgh August 2015



BioSharing şÝşÝߣs - Repository Fringe Edinburgh August 2015Peter McQuilton
Ěý
The document describes a web-based portal that registers, links, and allows discovery of biological standards and databases. It monitors the development and evolution of standards and their use in databases and adoption in data policies. Its mission is to provide support and guidance to researchers, developers, curators, journal publishers, and funders on choosing the appropriate standards and databases by registering, linking, and making discoverable relevant biological standards and databases.ELIXIR Webinar: BioSharing



ELIXIR Webinar: BioSharingPeter McQuilton
Ěý
This document provides an overview of BioSharing.org, a portal that monitors and curates standards, databases, and data policies to help inform and educate users. It summarizes key features including tracking over 225 content standards, 115 databases, and 554 data policies. The portal aims to help users understand how standards are used and make informed decisions on standards selection. It also links standards to training materials and allows custom collections and recommendations to be created.FAIRsharing and DataCite: Data Repository Selection- Criteria That Matter



FAIRsharing and DataCite: Data Repository Selection- Criteria That MatterSusanna-Assunta Sansone
Ěý
Through a collaboration with Datacite, FAIRsharing is working with a number of journal publishers (PLOS, Springer Nature, F1000, Wiley, Taylor and Francis, Elsevier, EMBO Press, eLife, GigaScience and Cambridge University Press) to identify a common set of criteria for selecting and recommending data repositories (and associated standards) that will be implemented in FAIRsharing. Details of this work and participants at https://osf.io/m2bceOSFair2017 Workshop | FAIRSharing



OSFair2017 Workshop | FAIRSharingOpen Science Fair
Ěý
Rafael Jimenez presents the FAIRSharing on behalf of Peter McQuilton, Susanna-Assunta Sansone & the FAIRsharing team | OSFair2017 Workshop
Workshop title: How FAIR friendly is your data catalogue?
Workshop overview:
This workshop will build upon the work planned by the EOSCpilot data interoperability task and the BlueBridge workshop held on April 3 at the RDA meeting. We will investigate common mechanisms for interoperation of data catalogues that preserve established community standards, norms and resources, while simplifying the process of being/becoming FAIR. Can we have a simple interoperability architecture based on a common set of metadata types? What are the minimum metadata requirements to expose FAIR data to EOSC services and EOSC users?
DAY 3 - PARALLEL SESSION 6 & 7BioSharing - RDA Plenary 6 - Metadata Standards Catalog WG and BioSharing WG ...



BioSharing - RDA Plenary 6 - Metadata Standards Catalog WG and BioSharing WG ...Peter McQuilton
Ěý
An introduction to the metadata landscape in the life sciences. Covering metadata standards, the databases that implement them and the policies that endorse/recommend both standards and databases in the life sciences."Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013



"Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013Susanna-Assunta Sansone
Ěý
Overview of the landscape of standards in life sciences for the NIH BD2K
"Frameworks for Community-Based Standards Efforts" workshop
September 25, 2013 - September 26, 2013
Co-Chairs: Susanna Sansone, PhD and David Kennedy PhD.
The overall goal of this workshop is to learn what has worked and what has not worked in community-based standards efforts. Participants will have experience in leading specific community based standards initiatives. Prior to the workshop, participants will be asked to address in writing answers to specific questions regarding formulating, conducting, and maintaining such efforts. This information will be used to facilitate focused and actionable discussion at the workshop. Issuance of a Request for Information soliciting comment from the broader community on some of the key issues addressed in the workshop is currently envisioned.
Contact: BD2Kworkshops@mail.nih.gov
Agenda: Frameworks for Community-Based Standards Efforts (PDF 40.7KB)
Participant List: Roster of Invited Participants (PDF 32KB)
Forum (Join the discussion): http://frameworks.prophpbb.com
Watch Live: http://videocast.nih.gov/summary.asp?live=13088 - See more at: http://bd2k.nih.gov/workshops.html#cbseOverview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014



Overview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014Susanna-Assunta Sansone
Ěý
What to know when planning for your data management strategy and preparing a data management statement for a research proposal for BBSRC DTP first year studentsThe BioSharing portal - linking journal and funder data policies to databases...



The BioSharing portal - linking journal and funder data policies to databases...Peter McQuilton
Ěý
A 20 minute talk on the BioSharing portal, focusing on our work to link journal and funder data policies to the databases and data standards that they recommend/endorse. This was presented as part of a session on data policies in the life sciences with representation from JISC and Springer Nature.FAIR data and NPG Scientific Data: RIKEN Yokohama, 25 June, 2014



FAIR data and NPG Scientific Data: RIKEN Yokohama, 25 June, 2014Susanna-Assunta Sansone
Ěý
The document summarizes a presentation about making scientific data FAIR (Findable, Accessible, Interoperable, Reusable). It discusses the concept of FAIR data and several of the presenter's related projects. Examples are provided of using standards like ISA-Tab to structure metadata and make datasets interoperable. The presentation outlines the presenter's roles in data capture, publication, and standards development efforts to promote FAIR data principles. Scientific Data, a new journal for peer-reviewed data descriptions, is introduced as a way to make datasets more discoverable and reusable.BioSharing - Mapping the landscape of Standards, Database and Data Policies i...



BioSharing - Mapping the landscape of Standards, Database and Data Policies i...Peter McQuilton
Ěý
A 20 minute talk on BioSharing, presented at the COST CHARME data standards conference in Warsaw, Poland, on June 21th 2016NIH iDASH meeting on data sharing - BioSharing, ISA and Scientific Data



NIH iDASH meeting on data sharing - BioSharing, ISA and Scientific DataSusanna-Assunta Sansone
Ěý
1) The document discusses Susanna-Assunta Sansone's roles and work related to promoting FAIR data standards and practices.
2) It highlights some of her leadership positions with organizations like BioSharing that work to map and promote standards.
3) The document also discusses Scientific Data, a peer-reviewed journal launched by Nature Publishing Group to publish detailed descriptions of scientifically valuable datasets to facilitate reuse.BioSharing, an ELIXIR Interoperability Platform resource



BioSharing, an ELIXIR Interoperability Platform resourcePeter McQuilton
Ěý
A 20 minute presentation given at the 9th RDA Plenary in Barcelona as part of the BioSharing WG - ELIXIR Bridging Force IG session. This presentation covers the basics of what BioSharing is, who it's for, and how it captures and connects information on data standards, databases and data policies from the life, biomedical and environmental sciences.The BioSharing portal - linking databases, data standards and policies in the...



The BioSharing portal - linking databases, data standards and policies in the...Peter McQuilton
Ěý
A 15 minute presentation for the Interest Group on Agricultural Data (IGAD) RDA pre-meeting meeting. Presented in Barcelona (ES) on Monday 3rd April, 2017.Managing Big Data - Berlin, July 9-10, 201.



Managing Big Data - Berlin, July 9-10, 201.Susanna-Assunta Sansone
Ěý
Susanna-Assunta Sansone is a data consultant and honorary academic editor who works on several projects related to making data FAIR (Findable, Accessible, Interoperable, Reusable). She is the associate director of Scientific Data, a peer-reviewed journal focused on publishing data descriptors to describe and provide access to scientifically valuable datasets. The goal of Scientific Data is to help promote open science and data reuse by publishing structured metadata and narratives about datasets alongside traditional research articles.Brief History of Public Health Systems and Services Research (PHSSR) 



Brief History of Public Health Systems and Services Research (PHSSR) National Coordinating Center for Public Health Services & Systems Research
Ěý
The document provides a history of Public Health Services and Systems Research (PHSSR) and the National Coordinating Center. It discusses how early work included developing data standards, funding research through mini-grants, and convening networking opportunities. It also summarizes how the coordinating center aims to advance the field through activities like establishing an endnote library, supporting practice-based research networks, and increasing communication and visibility of PHSSR through various channels. In closing, it reflects on lessons learned about the challenges of cross-sector coordination and promoting underrepresented areas of research like PHSSR."Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013



"Standards landscape" NIF Big Data 2 Knowledge (BD2K) Initiative, Sep, 2013Susanna-Assunta Sansone
Ěý
Overview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014



Overview to: BBSRC Oxford Doctoral Training Partnership - Dr Sansone - July 2014Susanna-Assunta Sansone
Ěý
Brief History of Public Health Systems and Services Research (PHSSR) 



Brief History of Public Health Systems and Services Research (PHSSR) National Coordinating Center for Public Health Services & Systems Research
Ěý
Recently uploaded (20)
Large Language Models (LLMs) part one.pptx



Large Language Models (LLMs) part one.pptxharmardir
Ěý
**The Rise and Impact of Large Language Models (LLMs)**
**Introduction**
In the rapidly evolving landscape of artificial intelligence (AI), one of the most groundbreaking advancements has been the development of Large Language Models (LLMs). These AI systems, trained on massive amounts of text data, have demonstrated remarkable capabilities in understanding, generating, and manipulating human language. LLMs have transformed industries, reshaped the way people interact with technology, and raised ethical concerns regarding their usage. This essay delves into the history, development, applications, challenges, and future of LLMs, providing a comprehensive understanding of their significance.
**Historical Background and Development**
The foundation of LLMs is built on decades of research in natural language processing (NLP) and machine learning (ML). Early language models were relatively simple and rule-based, relying on statistical methods to predict word sequences. However, the emergence of deep learning, particularly the introduction of neural networks, revolutionized NLP. The introduction of recurrent neural networks (RNNs) and long short-term memory (LSTM) networks in the late 1990s and early 2000s allowed for better sequential data processing.
The breakthrough moment for LLMs came with the development of Transformer architectures, introduced in the seminal 2017 paper "Attention Is All You Need" by Vaswani et al. The Transformer model enabled more efficient parallel processing and improved context understanding. This led to the creation of models like BERT (Bidirectional Encoder Representations from Transformers) and OpenAI’s GPT (Generative Pre-trained Transformer) series, which have since set new benchmarks in AI-driven text generation and comprehension.
**Core Mechanisms of LLMs**
LLMs rely on deep neural networks trained on extensive datasets comprising books, articles, websites, and other textual resources. The training process involves:
1. **Tokenization:** Breaking down text into smaller units (words, subwords, or characters) to be processed by the model.
2. **Pretraining:** The model learns general language patterns through unsupervised learning, predicting missing words or the next sequence in a text.
3. **Fine-tuning:** Adjusting the model for specific tasks, such as summarization, translation, or question-answering, using supervised learning.
4. **Inference:** The trained model generates text based on user input, leveraging probabilistic predictions to produce coherent responses.
Through these mechanisms, LLMs can perform a wide range of linguistic tasks with human-like proficiency.
**Applications of LLMs**
LLMs have found applications across various domains, including but not limited to:
1. **Content Generation:** LLMs assist in writing articles, blogs, poetry, and even code, helping content creators enhance productivity.
2. **Customer Support:** Chatbots and virtual assistants powered by LLMs provide automated yet cont19th Edition Of International Research Data Analysis Excellence Awards



19th Edition Of International Research Data Analysis Excellence Awardsdataanalysisconferen
Ěý
19th Edition Of International Research Data Analysis Excellence Awards
International Research Data Analysis Excellence Awards is the Researchers and Research organizations around the world in the motive of Encouraging and Honoring them for their Significant contributions & Achievements for the Advancement in their field of expertise. Researchers and scholars of all nationalities are eligible to receive ScienceFather Research Data Analysis Excellence Awards. Nominees are judged on past accomplishments, research excellence and outstanding academic achievements.
Place: San Francisco, United States
Visit Our Website: https://researchdataanalysis.com
Nomination Link: https://researchdataanalysis.com/award-nomination
Guide to Retrieval-Augmented Generation (RAG) and Contextual Augmented Genera...



Guide to Retrieval-Augmented Generation (RAG) and Contextual Augmented Genera...Doug Ortiz
Ěý
This document serves as a comprehensive guide to understanding Retrieval-Augmented Generation (RAG) and Contextual Augmented Generation (CAG). These innovative AI paradigms are reshaping content generation across various sectors. We will delve into their foundational principles, explore practical use cases, outline implementation strategies, and discuss performance measurement and common pitfalls.
MTC Supply Chain Management Strategy.pptx



MTC Supply Chain Management Strategy.pptxRakshit Porwal
Ěý
Strategic Cost Reduction in Medical Devices Supply Chain: Achieving Sustainable ProfitabilityThe Marketability of Rice Straw Yarn Among Selected Customers of Gantsilyo Guru



The Marketability of Rice Straw Yarn Among Selected Customers of Gantsilyo Gurukenyoncenteno12
Ěý
IMR PaperGlobalwits Global Standard Quotation 2025



Globalwits Global Standard Quotation 2025AvenGeorge1
Ěý
Globalwits GTIS6.0, the next-generation foreign trade big data analysis tool, covering 50%+ of global trade volume across 82 countries, 6 continents, and 255 trade databases. Designed to empower businesses with actionable insights, GTIS6.0 combines real-time analytics, AI-driven decision-making, and multi-dimensional intelligence to transform your global trade strategy.
Why Choose GTIS6.0?
Comprehensive Data Coverage
Track 100+ processing parameters, including buyer/supplier details, pricing trends, trade routes, and more.
Access real-time customs transaction data with 24/7 updates across 255 global trade databases.
Advanced Analytics & Visualization
Leverage RFM analysis, price interval tracking, and supply chain breakdown tools.
Generate customizable, business-grade reports with interactive charts (histograms, heatmaps, relational graphs).
Precision Targeting & Efficiency
Millisecond-level search speed powered by ElasticSearch and Hadoop technologies.
Identify high-potential markets and customers through multi-layered trade relationship mapping (up to 8 layers).
Seamless Integration & Resource Optimization
Bridge transaction data, public records, and business insights in one platform.
Utilize the Quality Supply-Demand Resource Sharing Platform to connect with verified buyers/suppliers worldwide.
Key Features:
Market Analysis: Visualize regional trade trends, product cycles, and competitor landscapes.
Customer Profiling: Evaluate buyer strength via procurement frequency, transaction value, and RFM metrics.
Decision-Maker Access: Retrieve official emails, LinkedIn profiles, and direct contacts to accelerate negotiations.
Dynamic Pricing Strategy: Analyze global price ranges and terms of trade for science-based quotations.
Real-Time Alerts: Monitor shifts in customer demand, partner changes, and supply chain risks.
SEO-Optimized Keywords:
Global Trade Data Analytics Tool, GTIS6.0 System, Foreign Trade Intelligence, Customs Data Insights, B2B Market Research, Supplier and Buyer Analysis, RFM Metrics, Trade Relationship Mapping, Globalwits Solutions, International Trade Optimization
Globalwits – Your Partner in Data-Driven Success
As the global standard for trade data, Globalwits bridges the gap between raw data and strategic decisions. Whether analyzing macro trends, profiling customers, or optimizing pricing, GTIS6.0 delivers unparalleled accuracy, speed, and depth.
Contact Us Today to Revolutionize Your Trade Strategy!
Director WhatsApp: +86 171 402 01001
Director Email: anyuchen@globalwits.comDesign Data Model Objects for Analytics, Activation, and AI



Design Data Model Objects for Analytics, Activation, and AIaaronmwinters
Ěý
Explore using industry-specific data standards to design data model objects in Data Cloud that can consolidate fragmented and multi-format data sources into a single view of the customer.
Design of the data model objects is a critical first step in setting up Data Cloud and will impact aspects of the implementation, including the data harmonization and mappings, as well as downstream automations and AI processing. This session will provide concrete examples of data standards in the education space and how to design a Data Cloud data model that will hold up over the long-term as new source systems and activation targets are added to the landscape. This will help architects and business analysts accelerate adoption of Data Cloud.Kaggle & Datathons: A Practical Guide to AI Competitions



Kaggle & Datathons: A Practical Guide to AI Competitionsrasheedsrq
Ěý
Kaggle & Datathons: A Practical Guide to AI Competitionsev3-software-tutorial-dc terminologia en ibgles.ppt



ev3-software-tutorial-dc terminologia en ibgles.ppthectormartinez817322
Ěý
en ingles esta prressetnacion es muy buena lasdla lkaskl siempreew aaslkd llasaslas as asasd lk aslk al kjal kj allksWhen Models Meet Data: From ancient science to todays Artificial Intelligence...



When Models Meet Data: From ancient science to todays Artificial Intelligence...ssuserbbbef4
Ěý
A presentation about Data and Machine LearningPlant Disease Prediction with Image Classification using CNN.pdf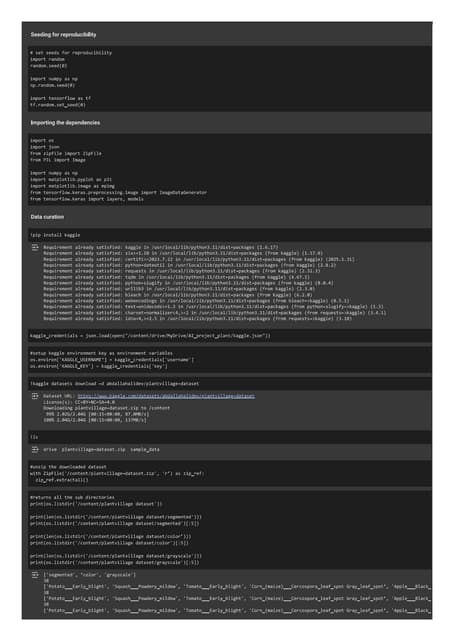



Plant Disease Prediction with Image Classification using CNN.pdfTheekshana Wanniarachchi
Ěý
Plant diseases pose a significant threat to agricultural productivity. Early detection and classification of plant diseases can help mitigate losses. This project focuses on building a Plant Disease Prediction system using Convolutional Neural Networks (CNNs). The system leverages NumPy, TensorFlow, and Streamlit to develop a model and deploy a web-based application. The final model is also containerized using Docker for efficient deployment.Cost sheet. with basics and formats of sheet



Cost sheet. with basics and formats of sheetsupreetk82004
Ěý
Cost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetCost sheet. with basics and formats of sheetUpdated Willow 2025 Media Deck_280225 Updated.pdf



Updated Willow 2025 Media Deck_280225 Updated.pdftangramcommunication
Ěý
Updated with the edits we last spoke BhavanaBEST MACHINE LEARNING INSTITUTE IS DICSITCOURSES



BEST MACHINE LEARNING INSTITUTE IS DICSITCOURSESgs5545791
Ěý
Machine learning is revolutionizing the way technology interacts with data, enabling systems to learn, adapt, and make intelligent decisions without human intervention. It plays a crucial role in various industries, from healthcare and finance to automation and artificial intelligence. If you want to build a successful career in this field, joining the Best Machine Learning Institute In Rohini is the perfect step. With expert-led training, hands-on projects, and industry-recognized certifications, you’ll gain the skills needed to thrive in the AI-driven world. If you are interested, then Enroll Fast – limited seats are available!
2024 Archive - Zsolt Nemeth web archivum



2024 Archive - Zsolt Nemeth web archivumZsolt Nemeth
Ěý
The 2024 Archive the Hollywood-Actresses.Com site new years collection.
Archivate blogging, link and clipart by Gigabajtos as Nemeth Zs. Selfpublisher active many organum or media-medium, written, comment, feed about Amazon books. Plus include 5.-Yearsbook (12.-NewsLetter).The truth behind the numbers: spotting statistical misuse.pptx



The truth behind the numbers: spotting statistical misuse.pptxandyprosser3
Ěý
As a producer of official statistics, being able to define what misinformation means in relation to data and statistics is so important to us.
For our sixth webinar, we explored how we handle statistical misuse especially in the media. We were also joined by speakers from the Office for Statistics Regulation (OSR) to explain how they play an important role in investigating and challenging the misuse of statistics across government.RDA WG proposal on connecting data policies, standards & databases in life sciences
- 1. Susanna-Assunta Sansone Associate Director, PI Rebecca Lawrence, Managing Director Simon Hodson, Executive Director Proposed RDA WG on Connecting data policies, standards and databases in the life sciences
- 2. PROPOSED WG: Connecting data policies, standards and databases in the life sciences Create a curated and cross-searchable registry of repositories, standards, and journal & funder policies. Help stakeholders to make informed decisions: • Journals on repositories accredited to the level required by their guidelines, but also meet the necessary standards. • Researchers on which journals meet their funder requirements and which repositories meet which journal standards. • Repositories on the requirements they need to meet to ensure researchers can use them to meet their funder policies and the policies of the journals they wish to publish in. • Funders on which journals and repositories meet their policies.
- 3. Growing number of reporting standards in life sciences + 130 Estimated + 150 Source:MIBBI, EQUATOR + 303 Source:BioPortal miame MIAPA MIRIAM MIQAS MIX MIGEN CIMR MIAPE MIASE MIQE MISFISHIE…. REMARK CONSORT MAGE-Tab GCDML SRAxml SOFT FASTA DICOM MzML SBRML SEDML… GELML ISA-Tab CML MITAB AAO CHEBI OBI PATO ENVO MOD BTO IDO… TEDDY PRO XAO DO VO To track provenance of the information and ensure richness of data and experimental metadata descriptions, to maximize reusability Databases, annotation, curation tools
- 4. But how much do we know about these standards
- 5. Increasing number of journals with differing data policies: Most major funders also now have open data policies but they all differ:
- 6. All Science WG: Global Service of Trusted Repositories and Services Life Sciences
- 7. * That has an MoU with *
- 8. Focus on life sciences: • Ensures manageable size of project • Pilot that is then extendable to other areas of science The plan: • Develop assessment criteria for usability and popularity of standards • Associate standards to data policies and databases • Assemble journal and funder policies re data storage • Make fully cross-searchable • Thereby help stakeholders make informed decisions Close interlinking with work of other relevant WGs (e.g. Global Service of Trusted Repositories WG; Metadata Standards Directory WG) to utilise outputs on the repository side will be very important






