Reverse transcription of hiv 1 ..
Download as PPTX, PDF1 like431 views
This is my another presentation in this platform. Hope you all can understand clearly through this presentation.
1 of 11
Download to read offline










Recommended
Transcription factors



Transcription factorsNehaliBuchade
╠²
This presentation is about the transcription machinery that is required for the transcription in eukaryotes. The comparison between the transcription factors involved in prokaryotes and eukaryotes. The initiation of transcription and how it helps in producing a mRNA. Ramachandran plot



Ramachandran plotRadhakrishna Gopala Pillai
╠²
The document discusses the Ramachandran plot, which shows statistically probable combinations of the phi and psi backbone torsion angles in proteins. It describes how these two angles describe rotations around bonds in the polypeptide backbone and influence protein folding. The plot reveals allowed and disallowed regions based on steric clashes between atoms at different angle combinations. Common structures like alpha helices and beta sheets correspond to allowed regions in the plot.Riboswitches and riboswitches mediated regulation



Riboswitches and riboswitches mediated regulationBhavikaKatariya
╠²
This document provides an overview of riboswitches and riboswitch-mediated gene regulation. It begins with an introduction that defines riboswitches as mRNA elements that bind metabolites or metal ions to regulate gene expression. The document then discusses the history and discovery of riboswitches, their structures, types of riboswitches including TPP, lysine, glycine and others, and their general mechanism of regulating gene expression through conformational changes. It also briefly discusses how riboswitches could potentially be targeted for novel antibiotic development and future challenges in understanding riboswitch function.Transcription



TranscriptionDr. A.D.Naveen Kumar
╠²
The document discusses transcription in prokaryotes and eukaryotes. In prokaryotes, RNA polymerase binds to promoter sequences and transcribes DNA into RNA through initiation, elongation, and termination. Transcription requires RNA polymerase and proceeds similarly in eukaryotes but involves multiple RNA polymerases and occurs in the nucleus. Eukaryotic transcription is more complex, utilizing regulatory sequences, transcription factors, and RNA processing to modify pre-mRNA into mature mRNA through splicing, capping, polyadenylation, and other modifications. Mutations can affect splicing and cause genetic disorders like beta-thalassemia.Rna silencing



Rna silencingUrooj Sabar
╠²
RNA silencing/RNAi involves the knock-down of genes through two types of small RNA molecules, miRNAs and siRNAs, that are involved in post-transcriptional and transcriptional gene silencing as an antiviral mechanism; short double-stranded RNAs are processed by the Dicer enzyme into siRNAs which are incorporated into the RISC complex to guide degradation of homologous mRNA targets; RNAi is an important endogenous gene regulatory mechanism and has applications in gene function analysis, gene therapy, and cancer treatment.Dna binding motiffs



Dna binding motiffsIndrajaDoradla
╠²
This document discusses different DNA binding motifs that allow proteins to interact with DNA without disrupting the hydrogen bonds between the DNA bases. It describes several conserved structural motifs common to many DNA binding proteins, including the helix-turn-helix motif, zinc finger domains, and leucine zipper domains. The helix-turn-helix motif contains two short alpha helices separated by a beta turn. Zinc finger domains use cysteine or histidine residues to coordinate a zinc ion, stabilizing their structure. Leucine zipper domains contain repeated leucine residues that allow dimerization of regulatory proteins.Eukaryotic transcription 2



Eukaryotic transcription 2Jilani Adnan
╠²
The central dogma describes how DNA is transcribed into RNA which is then translated into protein. Eukaryotic transcription involves RNA polymerases that transcribe DNA into RNA, with RNA polymerase II transcribing mRNA. The transcription process consists of initiation, elongation, and termination. It also involves transcription factors, chromatin remodeling, 5' capping, poly-A tail addition, and intron removal through splicing.Prokaryotic DNA replication



Prokaryotic DNA replicationMoumita Paul
╠²
DNA replication in prokaryotes begins with the unwinding of DNA at the origin of replication by enzymes like DnaA and DnaB helicase. This produces two replication forks that move in opposite directions. The leading strand is replicated continuously while the lagging strand is replicated discontinuously in short segments called Okazaki fragments. DNA polymerase III is the main enzyme that synthesizes new DNA. Replication terminates at the terminus region when the DnaB helicase is stopped by protein Tus bound to Ter sequences.DNA Binding Proteins.pptx



DNA Binding Proteins.pptxSNEHA AGRAWAL GUPTA
╠²
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA.It has two basic domains specifically recognises the target sequence. Histones are a special group of proteins found in the nuclei of eukaryotic cells responsible. Histones have five major classes : H1, H2A, H2B, H3 and H4.Gene regulation in eukaryotes



Gene regulation in eukaryotesIqra Wazir
╠²
Gene regulation in eukaryotes in a nutshell covering all the important stages of gene regulation in eukaryotes at transcriptional level, translation level and post-translational level.RNA PROCESSING



RNA PROCESSINGMOHDNADEEM68
╠²
RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA transcript is transformed into a mature messenger RNA. During splicing, introns are removed and exons are joined together.TRANSLATION



TRANSLATIONSurender Rawat
╠²
Messenger RNA carries genetic code from DNA and is translated by the ribosome into proteins. This involves transfer RNA molecules that associate amino acids with their codons. Translation begins with initiation factors recruiting the small ribosomal subunit to the start codon. Elongation then occurs through peptide bond formation catalyzed by the ribosome and translocation of transfer RNAs. Termination occurs when a stop codon is reached. Translation is highly conserved and essential for protein synthesis in all organisms.Protein dna interaction



Protein dna interactionPrasanthperceptron
╠²
The document discusses protein-DNA interactions and binding mechanisms. It covers several key points:
1) Specific binding of proteins to nucleic acids underlies gene expression processes like replication, repair, transcription and RNA metabolism.
2) Mechanisms of protein-DNA binding include the release of water molecules and ions, allowing the structures to conform and form hydrogen bonds and electrostatic interactions.
3) Recognition principles involve interactions with bases in the major/minor grooves, with certain amino acids preferring certain bases. Sequence-specific binding involves shape complementarity and electrostatic patches on the proteins.RNA SPLICING



RNA SPLICINGmanojjeya
╠²
RNA splicing is a process where introns are removed from precursor messenger RNA (pre-mRNA) and exons are joined together to produce mature mRNA. It occurs in the nucleus and is essential for eukaryotes to produce proteins. The spliceosome, a large complex of RNA and proteins, facilitates two transesterification reactions that remove introns and ligate exons. RNA splicing generates protein diversity through alternative splicing and is important for cellular functions and disease processes.Protein micro array



Protein micro arraykrupa sagar
╠²
Protein microarrays allow high-throughput analysis of protein interactions and functions. They consist of large numbers of capture proteins immobilized on a surface to which labeled probe molecules are added to detect reactions by fluorescence. There are analytical arrays to study protein binding properties and functional arrays containing full-length proteins to assay enzymatic activity and detect antibodies. Protein microarrays have applications in diagnostics, proteomics, analyzing protein interactions and functions, antibody characterization, and treatment development.Post-Translational Modifications



Post-Translational ModificationsAisha Kalsoom
╠²
The document discusses protein synthesis and post-translational modification. It describes how translation involves mRNA, ribosomes, tRNA, and release factors to synthesize proteins. The process involves initiation, elongation, and termination. After synthesis, the peptide undergoes folding, modification like phosphorylation, and can be transported to organelles. Post-translational modifications are important for diversity and regulating protein function, and involve processes like methylation, ubiquitination, and glycosylation. Diseases like atherosclerosis and fibrosis are related to disorders of collagen deposition and modification.Homology modelling



Homology modellingAyesha Choudhury
╠²
Prediction of the three dimensional structure of a given protein sequence i.e. target protein from the amino acid sequence of a homologous (template) protein for which an X-ray or NMR structure is available based on an alignment to one or more known protein structuresWobble hypothesis



Wobble hypothesissubhananthini jeyamurugan
╠²
- Crick proposed the "wobble hypothesis" to explain how more than one codon can direct the synthesis of a single amino acid, given there are fewer tRNAs than codons.
- The hypothesis suggests the third nucleotide in a codon is not as important in binding to the tRNA anticodon. The first two nucleotides specify the amino acid.
- At the wobble position, the third nucleotide in the codon can bind in non-standard ways ("wobble") to the first nucleotide in the anticodon, allowing a single tRNA to bind to multiple codons and explain the degeneracy of the genetic code.Gene expression



Gene expressionHina Zamir Noori
╠²
This document summarizes the process of gene expression in three main steps: transcription, post-transcriptional modification, and translation. It first defines a gene as a stretch of DNA that encodes information. During transcription, RNA polymerase produces messenger RNA from DNA. The mRNA then undergoes post-transcriptional modification like capping, splicing, and polyadenylation. The mature mRNA is transported to the cytoplasm for translation by ribosomes into proteins. Additional post-translational modifications can occur to proteins after translation. Gene expression is regulated at multiple levels including transcription, RNA processing, translation and protein degradation.Transcription in Eukaryotes



Transcription in EukaryotesRuchiRawal1
╠²
Transcription in eukaryotes is carried out by three RNA polymerases that synthesize different RNA molecules. RNA polymerase II initiates transcription through assembly of general transcription factors and a mediator complex at DNA promoter sequences. Initiation is followed by elongation and termination steps. Additional factors are required at each step to facilitate efficient transcription. The resulting RNA transcripts undergo processing including capping, polyadenylation, and export from the nucleus. Prokaryotic transcription differs in that it occurs in the cytoplasm and involves a single RNA polymerase, while eukaryotic transcription takes place in the nucleus and requires multiple RNA polymerases and transcription factors.RNA Processing



RNA ProcessingKartikey Rastogi
╠²
This document summarizes RNA processing. It discusses the key types of RNA - rRNA, tRNA, mRNA - and their roles. rRNA makes up ribosomes for protein synthesis. tRNA transfers amino acids during translation. mRNA carries genetic information from nucleus to cytoplasm. The document then explains that primary RNA transcripts undergo processing like 5' capping, 3' cleavage, polyadenylation, splicing, and editing to produce mature RNA molecules. Alternative splicing allows one gene to code for multiple proteins. In summary, this document provides an overview of RNA types and processing mechanisms that generate functional RNA from initial transcripts.Eukaryotic transcription



Eukaryotic transcriptionTanvi Potluri
╠²
Transcription in eukaryotes: A brief view
Transcription is the process by which single stranded RNA is synthesized by double stranded DNA. Transcription in eukaryotes and prokaryotes has many similarities while at the same time both showing their individual characteristics due to the differences in organization. RNA Polymerase (RNAP or RNA Pol) is different in prokaryotes and eukaryotes. Coupled transcription is seen in prokaryotes but not in Eukaryotes. In eukaryotes the pre-RNA should be spliced first to be translated.
In Eukaryotic transcription, synthesis of RNA occurs in the 3ŌĆÖŌåÆ5ŌĆÖ direction. The 3ŌĆÖ end is more reactive due to the hydroxide group. 5ŌĆÖ end containing phosphate groups meanwhile, is not very reactive when it comes to adding new nucleotides. In Eukaryotes, the whole genome is not transcribed at once. Only a part of the genome is transcribed which also acts as the first, principle stage of genetic regulation.
Eukaryotes have five nuclear polymerases:
ŌĆó RNA Polymerase I: This produces rRNA (23S, 5.8S, and 18S) which are the major components in a ribosome. This also produces pre-rRNA in yeasts.
ŌĆó RNA Polymerase II: Helps in the production of mRNA (messenger RNA), snRNA (small, nuclear RNA), miRNA. This is the most studied type and requires several transcription factors for its binding
ŌĆó RNA Polymerase III: This synthesizes tRNA (transfer RNA), 5S rRNA and other small RNAs required in the cytosol and nucleus.
ŌĆó RNA Polymerase IV: Synthesizes siRNA (small interfering RNA) in plants.
ŌĆó RNA Polymerase V: This is the least studied polymerase and synthesizes siRNA-directed heterochromatin in plants.
Eukaryotic transcription can be broadly divided into 4 stages:
ŌĆó Pre-Initiation
ŌĆó Initiation
ŌĆó Elongation
ŌĆó Termination
Transcription is an elaborate process which cells use to copy the genetic information stored in DNA into RNA. This pre-RNA is modified into mRNA before being transcribed to proteins. Transcription is the first step to utilizing the genetic information in a cell. Both Eukaryotes and Prokaryotes employ this process with the basic phases remaining the same. However eukaryotic transcription is more complex indicating the changes transcription has undergone towards perfection during evolution. SiRNA 



SiRNA Kashikant Yadav
╠²
Kashikant Yadav presented on siRNA (short interfering RNA). siRNA is 20-25 base pairs long, similar to miRNA, and operates in the RNA interference pathway by degrading mRNA with complementary sequences, preventing translation. There are three main methods of siRNA synthesis: chemical synthesis, in vitro transcription, and digestion of long dsRNA by RNAase III or Dicer. siRNA has significance for protecting against viruses, maintaining genome stability, and offers a new tool to specifically repress genes. Potential applications include testing gene function, target validation, pathway analysis, and developing siRNA therapeutics.Peptide mapping



Peptide mappingSaklecha Prachi
╠²
This document discusses peptide mapping, which involves enzymatically or chemically cleaving a protein into peptides and analyzing the peptides to identify the protein's primary structure. Peptide mapping is used to confirm identity, detect alterations, and ensure consistency for biopharmaceutical characterization. It involves four main steps - selectively cleaving peptide bonds, separating peptides chromatographically (often via RP-HPLC), analyzing and identifying peptides (usually via mass spectrometry), and comparing results to a reference standard to confirm identity. The document provides details on reagents, conditions, and parameters for each step of peptide mapping.Protein purification techniques



Protein purification techniquesNavaira Arif
╠²
The document discusses various techniques for purifying proteins. General steps in protein purification include selecting a protein source, homogenizing and solubilizing the proteins, and stabilizing the proteins. Specific techniques discussed include ammonium sulfate precipitation, dialysis, gel filtration chromatography, ion exchange chromatography, affinity chromatography, fast performance liquid chromatography, and gel electrophoresis. The goal of protein purification is to isolate a single protein of interest from other contaminating proteins in order to study its structure and function.Tryptophan operon



Tryptophan operondevadevi666
╠²
The tryptophan operon regulates the biosynthesis of tryptophan in E. coli through transcriptional attenuation and repression. It contains five genes encoding the enzymes needed to synthesize tryptophan. When tryptophan levels are high, the tryptophan repressor binds to the operator site, preventing transcription. Additionally, a regulatory region can form a terminator stem-loop structure to halt transcription if tryptophan tRNA levels are high during translation of the leader mRNA sequence. However, if tryptophan levels are low, the terminator structure does not form and transcription of the operon proceeds.Tm of DNA - melting temperature of DNA



Tm of DNA - melting temperature of DNAMandira bhosale
╠²
melting temperature of DNA
Chargaff's rule on DNA & RNA
denaturation renaturation
hyperchromism
hypochromism C value paradox



C value paradoxVishwasrao Naik Arts, Commerce And Baba Naik Science Mahavidyalaya, Shirala
╠²
This document discusses the C-Value Paradox, which is the observation that there is no correlation between the complexity of an organism and the amount of DNA (C-value) in its genome. The document provides examples showing that C-values, or the amount of DNA per haploid cell, can vary widely both within and across species, from 105 base pairs in mycoplasma to over 109 base pairs in mammals. While complexity tends to increase with higher C-values, exceptions exist, demonstrating there is no direct linear relationship between genome size and organism complexity. The term "C-value" refers to the haploid DNA content of a species.06. REVERSE TRANSCRIPTION, HIV AND ARVs.pptx



06. REVERSE TRANSCRIPTION, HIV AND ARVs.pptxMkindi Mkindi
╠²
This document discusses reverse transcription and antiretroviral therapy (ART) for HIV. It begins with an overview of reverse transcription, the process by which HIV converts its RNA genome into DNA. It then describes the mechanism of HIV reverse transcription in detail. The next section discusses how the synthesized HIV DNA is integrated into the host cell genome. The document concludes by covering the different classes of antiretroviral drugs, including nucleoside/nucleotide reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, protease inhibitors, integrase inhibitors, and entry inhibitors.Transcription; lgis



Transcription; lgisZahid Azeem
╠²
The document discusses DNA transcription in prokaryotes and eukaryotes. It defines transcription as the synthesis of RNA from a DNA template. In eukaryotes, transcription occurs in the nucleus and involves RNA polymerases I, II, and III. The process includes initiation, elongation, and termination. Eukaryotic transcription is more complex than prokaryotic transcription due to larger genome size, chromatin structure, and presence of regulatory elements. The primary transcript in eukaryotes undergoes processing before becoming a mature mRNA.More Related Content
What's hot (20)
DNA Binding Proteins.pptx



DNA Binding Proteins.pptxSNEHA AGRAWAL GUPTA
╠²
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA.It has two basic domains specifically recognises the target sequence. Histones are a special group of proteins found in the nuclei of eukaryotic cells responsible. Histones have five major classes : H1, H2A, H2B, H3 and H4.Gene regulation in eukaryotes



Gene regulation in eukaryotesIqra Wazir
╠²
Gene regulation in eukaryotes in a nutshell covering all the important stages of gene regulation in eukaryotes at transcriptional level, translation level and post-translational level.RNA PROCESSING



RNA PROCESSINGMOHDNADEEM68
╠²
RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA transcript is transformed into a mature messenger RNA. During splicing, introns are removed and exons are joined together.TRANSLATION



TRANSLATIONSurender Rawat
╠²
Messenger RNA carries genetic code from DNA and is translated by the ribosome into proteins. This involves transfer RNA molecules that associate amino acids with their codons. Translation begins with initiation factors recruiting the small ribosomal subunit to the start codon. Elongation then occurs through peptide bond formation catalyzed by the ribosome and translocation of transfer RNAs. Termination occurs when a stop codon is reached. Translation is highly conserved and essential for protein synthesis in all organisms.Protein dna interaction



Protein dna interactionPrasanthperceptron
╠²
The document discusses protein-DNA interactions and binding mechanisms. It covers several key points:
1) Specific binding of proteins to nucleic acids underlies gene expression processes like replication, repair, transcription and RNA metabolism.
2) Mechanisms of protein-DNA binding include the release of water molecules and ions, allowing the structures to conform and form hydrogen bonds and electrostatic interactions.
3) Recognition principles involve interactions with bases in the major/minor grooves, with certain amino acids preferring certain bases. Sequence-specific binding involves shape complementarity and electrostatic patches on the proteins.RNA SPLICING



RNA SPLICINGmanojjeya
╠²
RNA splicing is a process where introns are removed from precursor messenger RNA (pre-mRNA) and exons are joined together to produce mature mRNA. It occurs in the nucleus and is essential for eukaryotes to produce proteins. The spliceosome, a large complex of RNA and proteins, facilitates two transesterification reactions that remove introns and ligate exons. RNA splicing generates protein diversity through alternative splicing and is important for cellular functions and disease processes.Protein micro array



Protein micro arraykrupa sagar
╠²
Protein microarrays allow high-throughput analysis of protein interactions and functions. They consist of large numbers of capture proteins immobilized on a surface to which labeled probe molecules are added to detect reactions by fluorescence. There are analytical arrays to study protein binding properties and functional arrays containing full-length proteins to assay enzymatic activity and detect antibodies. Protein microarrays have applications in diagnostics, proteomics, analyzing protein interactions and functions, antibody characterization, and treatment development.Post-Translational Modifications



Post-Translational ModificationsAisha Kalsoom
╠²
The document discusses protein synthesis and post-translational modification. It describes how translation involves mRNA, ribosomes, tRNA, and release factors to synthesize proteins. The process involves initiation, elongation, and termination. After synthesis, the peptide undergoes folding, modification like phosphorylation, and can be transported to organelles. Post-translational modifications are important for diversity and regulating protein function, and involve processes like methylation, ubiquitination, and glycosylation. Diseases like atherosclerosis and fibrosis are related to disorders of collagen deposition and modification.Homology modelling



Homology modellingAyesha Choudhury
╠²
Prediction of the three dimensional structure of a given protein sequence i.e. target protein from the amino acid sequence of a homologous (template) protein for which an X-ray or NMR structure is available based on an alignment to one or more known protein structuresWobble hypothesis



Wobble hypothesissubhananthini jeyamurugan
╠²
- Crick proposed the "wobble hypothesis" to explain how more than one codon can direct the synthesis of a single amino acid, given there are fewer tRNAs than codons.
- The hypothesis suggests the third nucleotide in a codon is not as important in binding to the tRNA anticodon. The first two nucleotides specify the amino acid.
- At the wobble position, the third nucleotide in the codon can bind in non-standard ways ("wobble") to the first nucleotide in the anticodon, allowing a single tRNA to bind to multiple codons and explain the degeneracy of the genetic code.Gene expression



Gene expressionHina Zamir Noori
╠²
This document summarizes the process of gene expression in three main steps: transcription, post-transcriptional modification, and translation. It first defines a gene as a stretch of DNA that encodes information. During transcription, RNA polymerase produces messenger RNA from DNA. The mRNA then undergoes post-transcriptional modification like capping, splicing, and polyadenylation. The mature mRNA is transported to the cytoplasm for translation by ribosomes into proteins. Additional post-translational modifications can occur to proteins after translation. Gene expression is regulated at multiple levels including transcription, RNA processing, translation and protein degradation.Transcription in Eukaryotes



Transcription in EukaryotesRuchiRawal1
╠²
Transcription in eukaryotes is carried out by three RNA polymerases that synthesize different RNA molecules. RNA polymerase II initiates transcription through assembly of general transcription factors and a mediator complex at DNA promoter sequences. Initiation is followed by elongation and termination steps. Additional factors are required at each step to facilitate efficient transcription. The resulting RNA transcripts undergo processing including capping, polyadenylation, and export from the nucleus. Prokaryotic transcription differs in that it occurs in the cytoplasm and involves a single RNA polymerase, while eukaryotic transcription takes place in the nucleus and requires multiple RNA polymerases and transcription factors.RNA Processing



RNA ProcessingKartikey Rastogi
╠²
This document summarizes RNA processing. It discusses the key types of RNA - rRNA, tRNA, mRNA - and their roles. rRNA makes up ribosomes for protein synthesis. tRNA transfers amino acids during translation. mRNA carries genetic information from nucleus to cytoplasm. The document then explains that primary RNA transcripts undergo processing like 5' capping, 3' cleavage, polyadenylation, splicing, and editing to produce mature RNA molecules. Alternative splicing allows one gene to code for multiple proteins. In summary, this document provides an overview of RNA types and processing mechanisms that generate functional RNA from initial transcripts.Eukaryotic transcription



Eukaryotic transcriptionTanvi Potluri
╠²
Transcription in eukaryotes: A brief view
Transcription is the process by which single stranded RNA is synthesized by double stranded DNA. Transcription in eukaryotes and prokaryotes has many similarities while at the same time both showing their individual characteristics due to the differences in organization. RNA Polymerase (RNAP or RNA Pol) is different in prokaryotes and eukaryotes. Coupled transcription is seen in prokaryotes but not in Eukaryotes. In eukaryotes the pre-RNA should be spliced first to be translated.
In Eukaryotic transcription, synthesis of RNA occurs in the 3ŌĆÖŌåÆ5ŌĆÖ direction. The 3ŌĆÖ end is more reactive due to the hydroxide group. 5ŌĆÖ end containing phosphate groups meanwhile, is not very reactive when it comes to adding new nucleotides. In Eukaryotes, the whole genome is not transcribed at once. Only a part of the genome is transcribed which also acts as the first, principle stage of genetic regulation.
Eukaryotes have five nuclear polymerases:
ŌĆó RNA Polymerase I: This produces rRNA (23S, 5.8S, and 18S) which are the major components in a ribosome. This also produces pre-rRNA in yeasts.
ŌĆó RNA Polymerase II: Helps in the production of mRNA (messenger RNA), snRNA (small, nuclear RNA), miRNA. This is the most studied type and requires several transcription factors for its binding
ŌĆó RNA Polymerase III: This synthesizes tRNA (transfer RNA), 5S rRNA and other small RNAs required in the cytosol and nucleus.
ŌĆó RNA Polymerase IV: Synthesizes siRNA (small interfering RNA) in plants.
ŌĆó RNA Polymerase V: This is the least studied polymerase and synthesizes siRNA-directed heterochromatin in plants.
Eukaryotic transcription can be broadly divided into 4 stages:
ŌĆó Pre-Initiation
ŌĆó Initiation
ŌĆó Elongation
ŌĆó Termination
Transcription is an elaborate process which cells use to copy the genetic information stored in DNA into RNA. This pre-RNA is modified into mRNA before being transcribed to proteins. Transcription is the first step to utilizing the genetic information in a cell. Both Eukaryotes and Prokaryotes employ this process with the basic phases remaining the same. However eukaryotic transcription is more complex indicating the changes transcription has undergone towards perfection during evolution. SiRNA 



SiRNA Kashikant Yadav
╠²
Kashikant Yadav presented on siRNA (short interfering RNA). siRNA is 20-25 base pairs long, similar to miRNA, and operates in the RNA interference pathway by degrading mRNA with complementary sequences, preventing translation. There are three main methods of siRNA synthesis: chemical synthesis, in vitro transcription, and digestion of long dsRNA by RNAase III or Dicer. siRNA has significance for protecting against viruses, maintaining genome stability, and offers a new tool to specifically repress genes. Potential applications include testing gene function, target validation, pathway analysis, and developing siRNA therapeutics.Peptide mapping



Peptide mappingSaklecha Prachi
╠²
This document discusses peptide mapping, which involves enzymatically or chemically cleaving a protein into peptides and analyzing the peptides to identify the protein's primary structure. Peptide mapping is used to confirm identity, detect alterations, and ensure consistency for biopharmaceutical characterization. It involves four main steps - selectively cleaving peptide bonds, separating peptides chromatographically (often via RP-HPLC), analyzing and identifying peptides (usually via mass spectrometry), and comparing results to a reference standard to confirm identity. The document provides details on reagents, conditions, and parameters for each step of peptide mapping.Protein purification techniques



Protein purification techniquesNavaira Arif
╠²
The document discusses various techniques for purifying proteins. General steps in protein purification include selecting a protein source, homogenizing and solubilizing the proteins, and stabilizing the proteins. Specific techniques discussed include ammonium sulfate precipitation, dialysis, gel filtration chromatography, ion exchange chromatography, affinity chromatography, fast performance liquid chromatography, and gel electrophoresis. The goal of protein purification is to isolate a single protein of interest from other contaminating proteins in order to study its structure and function.Tryptophan operon



Tryptophan operondevadevi666
╠²
The tryptophan operon regulates the biosynthesis of tryptophan in E. coli through transcriptional attenuation and repression. It contains five genes encoding the enzymes needed to synthesize tryptophan. When tryptophan levels are high, the tryptophan repressor binds to the operator site, preventing transcription. Additionally, a regulatory region can form a terminator stem-loop structure to halt transcription if tryptophan tRNA levels are high during translation of the leader mRNA sequence. However, if tryptophan levels are low, the terminator structure does not form and transcription of the operon proceeds.Tm of DNA - melting temperature of DNA



Tm of DNA - melting temperature of DNAMandira bhosale
╠²
melting temperature of DNA
Chargaff's rule on DNA & RNA
denaturation renaturation
hyperchromism
hypochromism C value paradox



C value paradoxVishwasrao Naik Arts, Commerce And Baba Naik Science Mahavidyalaya, Shirala
╠²
This document discusses the C-Value Paradox, which is the observation that there is no correlation between the complexity of an organism and the amount of DNA (C-value) in its genome. The document provides examples showing that C-values, or the amount of DNA per haploid cell, can vary widely both within and across species, from 105 base pairs in mycoplasma to over 109 base pairs in mammals. While complexity tends to increase with higher C-values, exceptions exist, demonstrating there is no direct linear relationship between genome size and organism complexity. The term "C-value" refers to the haploid DNA content of a species.Similar to Reverse transcription of hiv 1 .. (20)
06. REVERSE TRANSCRIPTION, HIV AND ARVs.pptx



06. REVERSE TRANSCRIPTION, HIV AND ARVs.pptxMkindi Mkindi
╠²
This document discusses reverse transcription and antiretroviral therapy (ART) for HIV. It begins with an overview of reverse transcription, the process by which HIV converts its RNA genome into DNA. It then describes the mechanism of HIV reverse transcription in detail. The next section discusses how the synthesized HIV DNA is integrated into the host cell genome. The document concludes by covering the different classes of antiretroviral drugs, including nucleoside/nucleotide reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, protease inhibitors, integrase inhibitors, and entry inhibitors.Transcription; lgis



Transcription; lgisZahid Azeem
╠²
The document discusses DNA transcription in prokaryotes and eukaryotes. It defines transcription as the synthesis of RNA from a DNA template. In eukaryotes, transcription occurs in the nucleus and involves RNA polymerases I, II, and III. The process includes initiation, elongation, and termination. Eukaryotic transcription is more complex than prokaryotic transcription due to larger genome size, chromatin structure, and presence of regulatory elements. The primary transcript in eukaryotes undergoes processing before becoming a mature mRNA.RTPCRpresentationAmanNanavaty║▌║▌▀ŻShow.pdf



RTPCRpresentationAmanNanavaty║▌║▌▀ŻShow.pdfaman15nanavaty
╠²
This presentation talks on the mechanism of Reverse Transcription as well as RT-PCR. This presentation can be used to clearly understand the essential mechanism of RT-PCR, which has been gaining major analytical relevance. This presentation clearly elucidates the steps, advantages, inhibitors, required materials and disadvantages of RT-PCR in a simple and concise manner.Dna transcription by moun



Dna transcription by mounmounrafayel
╠²
The document discusses DNA transcription. It describes the process of transcription, including initiation, elongation, and termination. It discusses RNA polymerase, promoters, and post-transcriptional modifications. Key differences between prokaryotic and eukaryotic transcription are highlighted. The document also briefly mentions reverse transcription and some drugs that inhibit transcription.Transcription.ppt



Transcription.pptareenfakhoury1
╠²
Photosystem II captures and transfers energy.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
enter electron
transport chain
ŌĆō water molecules are
split
ŌĆō oxygen is released as
waste
ŌĆō hydrogen ions are
transported across
thylakoid membrane
4.1 Chemical Energy and ATP
ŌĆó Photosystem I captures energy and produces energycarrying molecules.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
are used to make
NADPH
ŌĆō NADPH is transferred
to light-independent
reactions
4.1 Chemical Energy and ATP
ŌĆó The light-dependent reactions produce ATP.
ŌĆō hydrogen ions flow through a channel in the thylakoid
membrane
ŌĆō ATP synthase attached to the channel makes ATP
4.1 Chemical Energy and ATP
ŌĆó Light-independent
reactions occur in the
stroma and use CO2
molecules.
The second stage of photosynthesis uses energy from
the first stage to make sugars.
4.1 Chemical Energy and ATP
ŌĆó A molecule of glucose is formed as it stores some of the
energy captured from sunlight.
ŌĆō carbon dioxide molecules enter the Calvin Photosystem II captures and transfers energy.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
enter electron
transport chain
ŌĆō water molecules are
split
ŌĆō oxygen is released as
waste
ŌĆō hydrogen ions are
transported across
thylakoid membrane
4.1 Chemical Energy and ATP
ŌĆó Photosystem I captures energy and produces energycarrying molecules.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
are used to make
NADPH
ŌĆō NADPH is transferred
to light-independent
reactions
4.1 Chemical Energy and ATP
ŌĆó The light-dependent reactions produce ATP.
ŌĆō hydrogen ions flow through a channel in the thylakoid
membrane
ŌĆō ATP synthase attached to the channel makes ATP
4.1 Chemical Energy and ATP
ŌĆó Light-independent
reactions occur in the
stroma and use CO2
molecules.
The second stage of photosynthesis uses energy from
the first stage to make sugars.
4.1 Chemical Energy and ATP
ŌĆó A molecule of glucose is formed as it stores some of the
energy captured from sunlight.
ŌĆō carbon dioxide molecules enter the Calvin Photosystem II captures and transfers energy.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
enter electron
transport chain
ŌĆō water molecules are
split
ŌĆō oxygen is released as
waste
ŌĆō hydrogen ions are
transported across
thylakoid membrane
4.1 Chemical Energy and ATP
ŌĆó Photosystem I captures energy and produces energycarrying molecules.
ŌĆō chlorophyll absorbs
energy from sunlight
ŌĆō energized electrons
are used to make
NADPH
ŌĆō NADPH is transferred
to light-independent
reactions
4.1 Chemical Energy and ATP
ŌĆó The light-dependent reactions produce ATP.
ŌĆō hydrogen ions flow through a channel in the thylakoid
membrane
ŌĆō ATP synthase attached to the channel makes ATP
4.1 Chemical Energy and ATP
ŌĆó Light-independent
reactions occur in the
stroma and use CO2
molecules.
The second stage of photosynthesis uses energy frvfTranscription.ppt



Transcription.pptKimEliakim1
╠²
Transcription is the process by which RNA polymerase converts DNA into RNA. It involves 3 stages: initiation, elongation, and termination. Initiation begins at promoter sequences on DNA and requires a sigma factor. During elongation, RNA polymerase proofreads as it synthesizes RNA to ensure accuracy. Termination occurs when the polymerase reaches termination sequences and releases the RNA transcript.Trabscription vishal



Trabscription vishalVISHAL SAXENA
╠²
dna transcription is a important topic for biology student. this presentation may be helpful for student of biology.it is useful for all types of courses as like M.Sc, B.Sc, 11th and 12th standard.
Transcription Dr Sohil



Transcription Dr SohilSohil Takodara
╠²
The document summarizes key differences between transcription in prokaryotes and eukaryotes. In prokaryotes, transcription and translation occur simultaneously in the cytoplasm using a single type of RNA polymerase. In eukaryotes, transcription occurs separately from translation in the nucleus using three distinct RNA polymerases and requires numerous transcription factors to initiate transcription. Eukaryotic transcription also involves more complex post-transcriptional processing including splicing of pre-mRNA and alternative splicing to generate multiple mRNA isoforms from a single gene.DNA Transcription (Pharmaceutical Biotechnology)



DNA Transcription (Pharmaceutical Biotechnology)Rikesh lal Shrestha
╠²
DNA Transcription in Prokaryotic and Eukaryotic Cell , Post Transcription modification, Reverse Transcription.TRANSCRIPTION.pptx



TRANSCRIPTION.pptxAliya Fathima Ilyas
╠²
ŌĆó Transcription machinery interacts with the template strand to produce an╠²mRNA╠²whose sequence resembles the coding strand.
ŌĆó Life on earth is said to have begun from self-replicating RNA since it is the only class of molecules capable of both catalysis and carrying genetic information.
ŌĆó Transcription maintains the link between these two molecules and allows cells to use a stable╠²nucleic acid╠²as the genetic material while retaining most of their╠²protein synthesis╠²machinery.
GENE EXPRESSION AND ITŌĆÖS REGULATION.pptx



GENE EXPRESSION AND ITŌĆÖS REGULATION.pptxyogesh532361
╠²
Gene expression is the process by which information from a gene is used to direct protein synthesis. It involves replication, transcription, and translation. Regulation of gene expression controls the amount and timing of gene products. Regulation occurs through transcription factors binding to regulatory elements near gene promoters to turn genes on and off. This precise control of gene expression is vital for cellular differentiation and adaptation.The flow of genetic information transcription



The flow of genetic information transcriptionLama K Banna
╠²
Genetics dentistry part 1 2017
Dr Mahmoud Serdah
https://dentist2015blog.wordpress.com/2017/06/11/genetics/Transcription.pptx



Transcription.pptxUdayShah40
╠²
Transcription is the process of copying DNA into RNA. In bacteria, RNA polymerase binds to promoter sequences and initiates transcription. It then elongates the RNA transcript by adding nucleotides complementary to the DNA template. Termination occurs via either Rho-dependent or Rho-independent mechanisms when the polymerase reaches a terminator sequence. In eukaryotes, RNA polymerase and transcription factors bind to promoter elements like the TATA box to initiate transcription. Termination is triggered by a polyadenylation signal that causes cleavage of the transcript.Transposons(jumping genes)



Transposons(jumping genes)Zaahir Salam
╠²
Transposons are segments of DNA that can move, or "jump", from one location in the genome to another. They were first discovered by Barbara McClintock in her studies of maize. Transposons make up over 50% of the human genome. There are two classes - Class I retrotransposons move via an RNA intermediate, while Class II transposons move directly from DNA to DNA. Transposition occurs through a "cut and paste" mechanism where the transposon is excised from one location and inserted into a new random site in the genome. While transposons can disrupt gene function, recent evidence suggests they may also help organisms adapt to environmental stress.Dna transcription



Dna transcriptionshree lakshmi
╠²
RNA is synthesized from DNA in a process called transcription. There are both similarities and differences between prokaryotic and eukaryotic transcription. In prokaryotes, transcription occurs in the cytoplasm, is carried out by a single type of RNA polymerase, and mRNA is transcribed directly from DNA. In eukaryotes, transcription occurs in the nucleus, utilizes three types of RNA polymerases, and produces hnRNA which is processed into mRNA. The key stages of transcription, initiation, elongation, and termination, occur through different mechanisms in prokaryotes and eukaryotes.DNA Transcription



DNA Transcriptionammara12
╠²
The document summarizes the process of transcription in cells. It defines transcription as the process by which RNA is synthesized using DNA as a template. It describes the basic structure of genes and the roles of different types of RNA including mRNA, tRNA, rRNA and snRNA. The transcription process involves RNA polymerase binding to the promoter region and synthesizing RNA along the DNA template, adding complementary bases. Elongation continues until a terminator sequence is reached, at which point transcription ends and the RNA transcript is released.Transcription in prokaryotes and eukaryotes.pdf



Transcription in prokaryotes and eukaryotes.pdfssuser880f82
╠²
1. Transcription is the process of copying DNA into mRNA, which carries genetic information from DNA in the nucleus to the cytoplasm for protein production. RNA polymerase copies one DNA strand into a complementary mRNA strand.
2. In prokaryotes, RNA polymerase initiates transcription by binding to promoter sequences upstream of genes. It then elongates the mRNA as it moves along the DNA template. Termination occurs via rho-dependent or rho-independent signals.
3. In eukaryotes, transcription requires transcription factors to recruit RNA polymerase to the promoter. The initial pre-mRNA product undergoes processing before becoming a mature mRNA for translation.Transcription (Eukaryotic and prokaryotic )



Transcription (Eukaryotic and prokaryotic )MujeebUrRehman70
╠²
This document provides an overview of transcription in prokaryotes and eukaryotes. It describes that transcription is the first step of gene expression where RNA is synthesized from a DNA template. In prokaryotes, transcription occurs in the cytoplasm and is carried out by RNA polymerase, while in eukaryotes it occurs in the nucleus and requires transcription factors. The process involves initiation, elongation, and termination stages. In prokaryotes, RNA polymerase binds directly to promoter sequences, while in eukaryotes transcription factors are needed to recruit RNA polymerase. The document compares the key differences between prokaryotic and eukaryotic transcription.Recently uploaded (20)
Functional Muscle Testing of Facial Muscles.pdf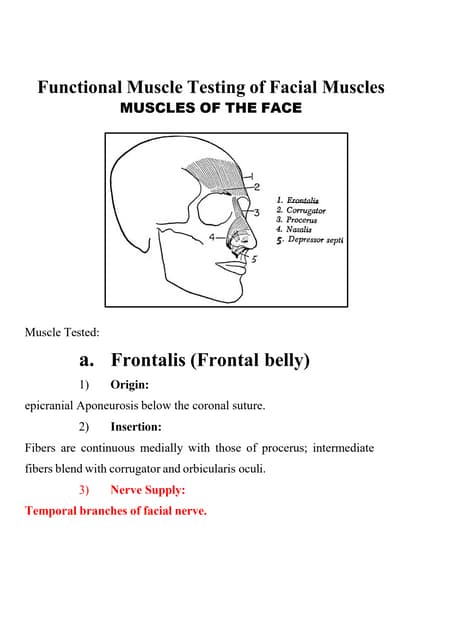



Functional Muscle Testing of Facial Muscles.pdfSamarHosni3
╠²
Functional Muscle Testing of Facial Muscles.pdfMastering Soft Tissue Therapy & Sports Taping



Mastering Soft Tissue Therapy & Sports TapingKusal Goonewardena
╠²
Mastering Soft Tissue Therapy & Sports Taping: Pathway to Sports Medicine Excellence
This presentation was delivered in Colombo, Sri Lanka, at the Institute of Sports Medicine to an audience of sports physiotherapists, exercise scientists, athletic trainers, and healthcare professionals. Led by Kusal Goonewardena (PhD Candidate - Muscle Fatigue, APA Titled Sports & Exercise Physiotherapist) and Gayath Jayasinghe (Sports Scientist), the session provided comprehensive training on soft tissue assessment, treatment techniques, and essential sports taping methods.
Key topics covered:
Ō£ģ Soft Tissue Therapy ŌĆō The science behind muscle, fascia, and joint assessment for optimal treatment outcomes.
Ō£ģ Sports Taping Techniques ŌĆō Practical applications for injury prevention and rehabilitation, including ankle, knee, shoulder, thoracic, and cervical spine taping.
Ō£ģ Sports Trainer Level 1 Course by Sports Medicine Australia ŌĆō A gateway to professional development, career opportunities, and working in Australia.
This training mirrors the Elite Akademy Sports Medicine standards, ensuring evidence-based approaches to injury management and athlete care.
If you are a sports professional looking to enhance your clinical skills and open doors to global opportunities, this presentation is for you.How to Configure Recurring Revenue in Odoo 17 CRM



How to Configure Recurring Revenue in Odoo 17 CRMCeline George
╠²
This slide will represent how to configure Recurring revenue. Recurring revenue are the income generated at a particular interval. Typically, the interval can be monthly, yearly, or we can customize the intervals for a product or service based on its subscription or contract. Hannah Borhan and Pietro Gagliardi OECD present 'From classroom to community ...



Hannah Borhan and Pietro Gagliardi OECD present 'From classroom to community ...EduSkills OECD
╠²
Hannah Borhan, Research Assistant, OECD Education and Skills Directorate and Pietro Gagliardi, Policy Analyst, OECD Public Governance Directorate present at the OECD webinar 'From classroom to community engagement: Promoting active citizenship among young people" on 25 February 2025. You can find the recording of the webinar on the website https://oecdedutoday.com/webinars/
Dr. Ansari Khurshid Ahmed- Factors affecting Validity of a Test.pptx



Dr. Ansari Khurshid Ahmed- Factors affecting Validity of a Test.pptxKhurshid Ahmed Ansari
╠²
Validity is an important characteristic of a test. A test having low validity is of little use. Validity is the accuracy with which a test measures whatever it is supposed to measure. Validity can be low, moderate or high. There are many factors which affect the validity of a test. If these factors are controlled, then the validity of the test can be maintained to a high level. In the power point presentation, factors affecting validity are discussed with the help of concrete examples.Dot NET Core Interview Questions PDF By ScholarHat



Dot NET Core Interview Questions PDF By ScholarHatScholarhat
╠²
Dot NET Core Interview Questions PDF By ScholarHatRRB ALP CBT 2 RAC Question Paper MCQ (Railway Assistant Loco Pilot)



RRB ALP CBT 2 RAC Question Paper MCQ (Railway Assistant Loco Pilot)SONU HEETSON
╠²
RRB ALP CBT 2 RAC Question Paper MCQ PDF Free Download. Railway Assistant Loco Pilot Mechanic Refrigeration and Air Conditioning Important Questions.How to Configure Proforma Invoice in Odoo 18 Sales



How to Configure Proforma Invoice in Odoo 18 SalesCeline George
╠²
In this slide, weŌĆÖll discuss on how to configure proforma invoice in Odoo 18 Sales module. A proforma invoice is a preliminary invoice that serves as a commercial document issued by a seller to a buyer.RRB ALP CBT 2 Mechanic Motor Vehicle Question Paper (MMV Exam MCQ)



RRB ALP CBT 2 Mechanic Motor Vehicle Question Paper (MMV Exam MCQ)SONU HEETSON
╠²
RRB ALP CBT 2 Mechanic Motor Vehicle Question Paper. MMV MCQ PDF Free Download for Railway Assistant Loco Pilot Exam.How to Configure Deliver Content by Email in Odoo 18 Sales



How to Configure Deliver Content by Email in Odoo 18 SalesCeline George
╠²
In this slide, weŌĆÖll discuss on how to configure proforma invoice in Odoo 18 Sales module. A proforma invoice is a preliminary invoice that serves as a commercial document issued by a seller to a buyer.Interim Guidelines for PMES-DM-17-2025-PPT.pptx



Interim Guidelines for PMES-DM-17-2025-PPT.pptxsirjeromemanansala
╠²
This is the latest issuance on PMES as replacement of RPMS. Kindly message me to gain full access of the presentation. Comprehensive Guide to Antibiotics & Beta-Lactam Antibiotics.pptx



Comprehensive Guide to Antibiotics & Beta-Lactam Antibiotics.pptxSamruddhi Khonde
╠²
¤ōó Comprehensive Guide to Antibiotics & Beta-Lactam Antibiotics
¤ö¼ Antibiotics have revolutionized medicine, playing a crucial role in combating bacterial infections. Among them, Beta-Lactam antibiotics remain the most widely used class due to their effectiveness against Gram-positive and Gram-negative bacteria. This guide provides a detailed overview of their history, classification, chemical structures, mode of action, resistance mechanisms, SAR, and clinical applications.
¤ōī What YouŌĆÖll Learn in This Presentation
Ō£ģ History & Evolution of Antibiotics
Ō£ģ Cell Wall Structure of Gram-Positive & Gram-Negative Bacteria
Ō£ģ Beta-Lactam Antibiotics: Classification & Subtypes
Ō£ģ Penicillins, Cephalosporins, Carbapenems & Monobactams
Ō£ģ Mode of Action (MOA) & Structure-Activity Relationship (SAR)
Ō£ģ Beta-Lactamase Inhibitors & Resistance Mechanisms
Ō£ģ Clinical Applications & Challenges.
¤ÜĆ Why You Should Check This Out?
Essential for pharmacy, medical & life sciences students.
Provides insights into antibiotic resistance & pharmaceutical trends.
Useful for healthcare professionals & researchers in drug discovery.
¤æē Swipe through & explore the world of antibiotics today!
¤öö Like, Share & Follow for more in-depth pharma insights!NUTRITIONAL ASSESSMENT AND EDUCATION - 5TH SEM.pdf



NUTRITIONAL ASSESSMENT AND EDUCATION - 5TH SEM.pdfDolisha Warbi
╠²
NUTRITIONAL ASSESSMENT AND EDUCATION, Introduction, definition, types - macronutrient and micronutrient, food pyramid, meal planning, nutritional assessment of individual, family and community by using appropriate method, nutrition education, nutritional rehabilitation, nutritional deficiency disorder, law/policies regarding nutrition in India, food hygiene, food fortification, food handling and storage, food preservation, food preparation, food purchase, food consumption, food borne diseases, food poisoningReverse transcription of hiv 1 ..
- 2. Abstract:- ŌĆó Reverse transcription and integration are the defining features of the Retroviridae; the common name ŌĆ£retrovirusŌĆØ derives from the fact that these viruses use a virally encoded enzyme, reverse transcriptase (RT), to convert their RNA genomes into DNA. Reverse transcription is an essential step in retroviral replication.
- 3. The Process of Reverse Transcription:- ŌĆó When a mature HIV-1 virion infects a susceptible target cell, interactions of the envelope glycoprotein with the coreceptors on the surface of the cell brings about a fusion of the membranes of the host cell and the virion . This fusion introduces the contents of the virion into the cytoplasm of the cell, setting the stage for reverse transcription.
- 4. ŌĆó Reverse transcription initiates from an RNA primer provided by a specific cellular tRNA (tRNA Lys3). The primer hybridizes to the primer Binding Site (PBS) of the genomic RNA. ŌĆó Reverse transcriptase catalyzes RNA dependent DNA synthesis. Synthesis of the minus-strand DNA generates an RNA-DNA hybrid. Rnase H activity of reverse transcriptase degrades the RNA strand, leaving the nascent minus-strand DNA single stranded. ŌĆó The newly synthesized minus-strand DNA hybridizes with the terminal R-region of the 3ŌĆÖ ŌĆō end of the viral RNA ( Called the first jump or minus-strand transfer).
- 5. ŌĆó After the nascent DNA hybridizes to R, minus--strand DNA synthesis can continue along the viral RNA. As DNA synthesis proceeds, RNase H degrades the RNA strand.
- 6. ŌĆó There is a Purine-rich sequence ( called the polypurine tract or PPT) near the 3ŌĆÖ end of the viral RNA which is relatively resistant to RNase H cleavage, and it serves as the primer for plus -- strand DNA synthesis. ŌĆó Plus-strand DNA synthesis proceeds until reverse transcriptase starts to copy the tRNA primer. Once the 3ŌĆÖ end of the tRNA has been reverse transcribed , an RNA/DNA hybrid that is a substrate for RNase H is created.
- 7. ŌĆó The tRNA associated with the nascent viral DNA is then removed by the RNase H activity. This may facilitate annealing to the PBS complement on the minus-strand DNA, providing the complementarity for the Second jump or Plus-strand transfer. ŌĆó Once this Second transfer happens, both the minus ŌĆō and plus ŌĆō DNA strands are extended until the entire DNA is double stranded and creating a DNA that has the same sequences at both ends (these repeats are called the Long Terminal Repeats or LTRs).
- 9. Why Left hand LTR and Right hand LTR is necessary ? ŌĆó RNA polymerase that initate transcription with in the Left hand LTR can dislodge transcription factor bound to the Right hand LTR as they passed through that region on the proviral DNA inactivating itŌĆÖs ability to initiate for transcription. This is called Promotar occlusion. This concept is supported by the observation that mutation or deletion of the Left hand LTR can lead to the initiation of the transcription in the Right LTR resulting in transcription of the cellular genes downstream of the proviral DNA.
- 10. ŌĆó The cellular cleavage and poly adenylation machinery recognizes the highly conserved AAUAAA poly adenylation signal in RNA.
- 11. THANK YOU