Scalable Text Mining
- 1. Scalable Text Mining Jee-Hyub Kim Text-Mining Pipeline Builder Literature Services Team 2 Feb 2016
- 3. Contents â Text-Mining Pipeline Crisis â Session 1: Build Your Own Pipeline â Session 2: Build Your Own Dictionary â Wrap Up
- 4. Use case Semantic type Dictionary type Document type Section Metadata Delivery method OpenAIRE accession numbers pattern (e.g, [0-9][A- Za-z0-9]{3}) patents Title, Claim, Description, Abstract, Figure, Table Pubyear, IPCR summary table ERC grant identifiers pattern articles Acknowledgements search index CTTV gene, disease term (e.g., IBD) articles, abstracts json ELIXIR-EXCELERTAE resource names term articles summary table 1000 Genomes cell line names pattern articles !Acknowledgements REST API Wikipedia accession numbers pattern wikipages summary table KEW Garden species names (muitilingual) term articles summary table ChEMBL resource name term articles Author, Journal summary table Ensembl genomic range pattern articles summary table A long list of requests
- 5. Scalable Text Mining â For the last few years, weâre having a pipeline crisis! â A long list of requests and our slow responses â Makes you unhappy. â Even worse, itâs a long tail! â Never the same pipeline used for each request. â Every time, we have to build a new pipeline. â We need a new approach to solve this crisis.
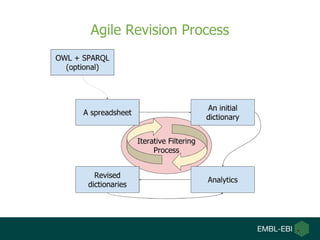
- 6. Objective â We want to build a LEGO-like platform that helps you to build your own text-mining pipeline and your own dictionary.
- 7. A Key Block: Dictionary-Based Tagger â Role: To identify names (e.g., proteins, species, accession numbers, etc.) â Dictionary-based approach for mining names. â Simple â Readable â Interactive â Building a dictionary is a VERY iterative process â 20% for building an initial dictionary and the rest for refining it. â Good dictionaries are a key for text-mining success stories.
- 9. Session 1 Build Your Own Pipeline As âĶ, I want a pipeline to do ...
- 10. Pipeline Stories â CTTV â As a researcher, I want to find articles with supporting evidence from drug discovery â ERC â As a funder, I want to funded articles more searchable. â ELIXIR-EXCELERATE â As a resource manager, I want to know impacts of resources.
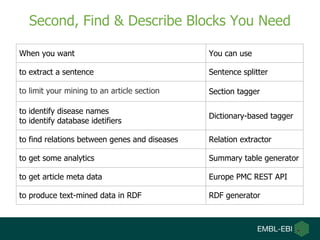
- 11. Second, Find & Describe Blocks You Need When you want You can use to extract a sentence Sentence splitter to limit your mining to an article section Section tagger to identify disease names to identify database idetifiers Dictionary-based tagger to find relations between genes and diseases Relation extractor to get some analytics Summary table generator to get article meta data Europe PMC REST API to produce text-mined data in RDF RDF generator
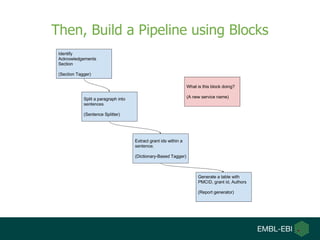
- 12. Then, Build a Pipeline using Blocks
- 13. Session 2 Build Your Own Dictionary Designing filtering rules
- 14. How to Revise a Dictionary? â We want to build an expressive language for filtering. â Global filtering rule â A length of term > 2 â Case sensitive â Per-entry filtering rule â A term should be tagged when it is mentioned in Methods section. â A pattern should be tagged when it follows a term âomimâ â Blacklist: e.g., stop words
- 15. Per-Entry Rules â A spreadsheet per entry â Definitions â Context: should (not) be after a tem. â Section: should (not) be mentioned a section. â URI: check if http://www.ebi.ac. uk/efo/EFO_0001997 exists Entry information Filtering rules Term/Pattern Entry ID DB Context Section URI Pattern HG[0-9]{5} 1000 genomes ! (grant|fun d) !ACK Term basal cell EFO_0001997 efo Methods Yes
- 16. Analytics â Summary table â Top 100 frequent terms PMCID Term ID Frequency PMCID4698870 Nutlin-3 ChEBI:46742 16 PMCID4698870 cell cycle arrests GO:0007050 6 Top Name Document Freq. Collection Freq. 1 protein 678,987 1,823,783 2 water 563,234 1,233,332
- 17. Spreadsheet for Filtering Rules http://tinyurl.com/zlwbx2y
- 18. Wrap Up â What is your pipeline story? â Have you managed to create your own dictionary? â What service blocks are missing? â What should be the interfaces? â How should we deliver?




![Use case Semantic type
Dictionary
type
Document
type
Section Metadata Delivery method
OpenAIRE
accession
numbers
pattern
(e.g, [0-9][A-
Za-z0-9]{3})
patents
Title, Claim,
Description,
Abstract, Figure,
Table
Pubyear,
IPCR
summary table
ERC grant identifiers pattern articles Acknowledgements search index
CTTV gene, disease
term
(e.g., IBD)
articles,
abstracts
json
ELIXIR-EXCELERTAE resource names term articles summary table
1000 Genomes cell line names pattern articles !Acknowledgements REST API
Wikipedia
accession
numbers
pattern wikipages summary table
KEW Garden
species names
(muitilingual)
term articles summary table
ChEMBL resource name term articles
Author,
Journal
summary table
Ensembl genomic range pattern articles summary table
A long list of requests](https://image.slidesharecdn.com/scalabletextminingv32-160224100559/85/Scalable-Text-Mining-4-320.jpg)







![Per-Entry Rules
â A spreadsheet per entry
â Definitions
â Context: should (not) be after a tem.
â Section: should (not) be mentioned a section.
â URI: check if http://www.ebi.ac.
uk/efo/EFO_0001997 exists
Entry information Filtering rules
Term/Pattern Entry ID DB Context Section URI
Pattern HG[0-9]{5}
1000
genomes
!
(grant|fun
d)
!ACK
Term basal cell EFO_0001997 efo Methods Yes](https://image.slidesharecdn.com/scalabletextminingv32-160224100559/85/Scalable-Text-Mining-15-320.jpg)


