Splendid: SPARQL Endpoint Federation Exploiting VOID Descriptions
- 1. Institute for Web Science and Technologies University of Koblenz Ō¢¬ Landau, Germany SPLENDID: SPARQL Endpoint Federation Exploiting VOID Descriptions Olaf G├Črlitz, Steffen Staab
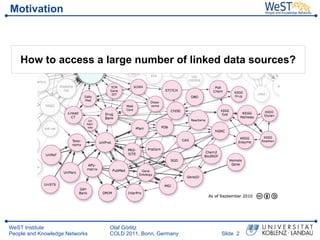
- 2. Motivation How to access a large number of linked data sources? WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 2
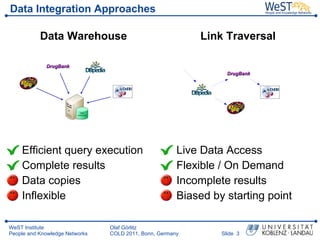
- 3. Data Integration Approaches Data Warehouse Link Traversal ’üĘ Efficient query execution ’üĘ Live Data Access ’üĘ Complete results ’üĘ Flexible / On Demand ’üĘ Data copies ’üĘ Incomplete results ’üĘ Inflexible ’üĘ Biased by starting point WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 3
- 4. Our Approach Data Federation Live data access Flexible source integration Effective query planning Complete results Hypothesis: Efficient query federation is possible using core Semantic Web technology (i.e. SPARQL endpoints, VoiD descriptions) WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 4
- 5. VoiD: ŌĆ×Vocabulary of Interlinked DatasetsŌĆ£ } General Information } Basic statistics triples = 732744 } Type statistics chebi:Compound = 50477 } Predicate statistics bio:formula = 39555 WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 5
- 6. Distributed Query Processing Contribution: Apply Best Practices of RDBMS for RDF Federation http://code.google.com/p/rdffederator/ WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 6
- 7. Query Example Which drugs are categorized as micronutrients? SELECT╠²?drug╠²?title╠²WHERE╠²{ ╠²╠²?drug╠²drugbank:drugCategory╠²category:micronutrient╠². ╠²╠²?drug╠²drugbank:casRegistryNumber╠²?id╠². ╠²╠²?keggDrug╠²rdf:type╠²kegg:Drug╠². ╠²╠²?keggDrug╠²bio2rdf:xRef╠²?id╠². ╠²╠²?keggDrug╠²purl:title╠²?title╠².╠²} } WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 7
- 8. Query Processing Source Selection Join Optimization Query Execution SELECT╠²?drug╠²?title╠²WHERE╠²{ ╠²╠²?drug╠²drugbank:drugCategory╠²category:micronutrient╠². ╠²╠²?drug╠²drugbank:casRegistryNumber╠²?id╠². ╠²╠²?keggDrug╠²rdf:type╠²kegg:Drug╠². ╠²╠²?keggDrug╠²bio2rdf:xRef╠²?id╠². ╠²╠²?keggDrug╠²purl:title╠²?title╠².╠²} } WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 8
- 9. Query Processing Source Selection Join Optimization Query Execution 1. Step: Index-based source mapping SELECT╠²?drug╠²?title╠²WHERE╠²{ ╠²╠²?drug╠²drugbank:drugCategory╠²category:micronutrient╠². ŌåÆ drugbank ╠²╠²?drug╠²drugbank:casRegistryNumber╠²?id╠². ŌåÆ drugbank ╠²╠²?keggDrug╠²rdf:type╠²kegg:Drug╠². ŌåÆ kegg ╠²╠²?keggDrug╠²bio2rdf:xRef╠²?id╠². ŌåÆ kegg ╠²╠²?keggDrug╠²purl:title╠²?title╠².╠²} ŌåÆ kegg, dbpedia, Chebi } predicate-index type-index drugbank:drugCategory ŌåÆ drugbank kegg:Drug ŌåÆ kegg WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 9
- 10. Query Processing Source Selection Join Optimization Query Execution 2. Step: Refinement with ASK Queries SELECT╠²?drug╠²?title╠²WHERE╠²{ ╠²╠²?drug╠²drugbank:drugCategory╠²category:micronutrient╠². ╠²╠²?drug╠²drugbank:casRegistryNumber╠²?id╠². ╠²╠²?keggDrug╠²rdf:type╠²kegg:Drug╠². ╠²╠²?keggDrug╠²bio2rdf:xRef╠²?id╠². ╠²╠²?keggDrug╠²purl:title╠²?title╠².╠²} } No index for subject / object values WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 10
- 11. Query Processing Source Selection Join Optimization Query Execution 3. Step: Grouping Triple Patterns SELECT╠²?drug╠²?title╠²WHERE╠²{ ╠²╠²?drug╠²drugbank:drugCategory╠²category:micronutrient╠². ╠²╠²?drug╠²drugbank:casRegistryNumber╠²?id╠². } drugbank ╠²╠²?keggDrug╠²rdf:type╠²kegg:Drug╠². ╠²╠²?keggDrug╠²bio2rdf:xRef╠²?id╠². } kegg ╠²╠²?keggDrug╠²purl:title╠²?title╠².╠²} } kegg, dbpedia, Chebi } + grouping sameAs patterns WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 11
- 12. Join Order Optimization Source Selection Join Optimization Query Execution Dynamic Programming with statistics-based cost estimation bind join / hash join WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 12
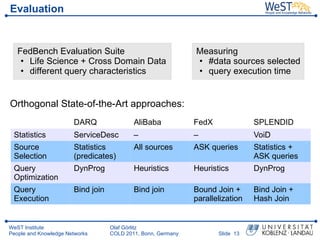
- 13. Evaluation FedBench Evaluation Suite Measuring ŌĆó Life Science + Cross Domain Data ŌĆó #data sources selected ŌĆó different query characteristics ŌĆó query execution time Orthogonal State-of-the-Art approaches: DARQ AliBaba FedX SPLENDID Statistics ServiceDesc ŌĆō ŌĆō VoiD Source Statistics All sources ASK queries Statistics + Selection (predicates) ASK queries Query DynProg Heuristics Heuristics DynProg Optimization Query Bind join Bind join Bound Join + Bind Join + Execution parallelization Hash Join WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 13
- 14. Evaluation: Source Selection Source Selection Join Optimization Query Execution owl:sameAs rdf:type WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 14
- 15. Evaluation: Query Optimization Source Selection Join Optimization Query Execution WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 15
- 16. Conclusion Publish more VoiD description! VoiD-based query federation is efficient What next? ’üĘ Combination with FedX ’üĘ Improving estimation and cost model ’üĘ Integrating SPARQL 1.1 features WeST Institute Olaf G├Črlitz People and Knowledge Networks COLD 2011, Bonn, Germany ║▌║▌▀Ż 16
Editor's Notes
- #3: Pre-selected linked datasets Transparent query federation