Transposable elements
Download as PPTX, PDF1 like1,879 views
Barbara McClintock discovered transposons in the 1940s through her genetic experiments with corn. Transposons are DNA sequences that can move and insert themselves in new locations in the genome. Over 50% of the human genome consists of transposons, particularly retrotransposons, which can insert via reverse transcription. Several marker techniques have been developed that exploit the insertion polymorphisms of retrotransposons, including S-SAP, IRAP, REMAP and RBIP. These markers allow for studies of genetic variation, gene mapping and phylogenetics.
1 of 22
Downloaded 24 times




















Recommended
Bacterial transposable elements



Bacterial transposable elementsTejaswini Petkar
Ėý
Transposable elements are DNA sequences that can change their position within a genome. They are common in bacteria and include insertion sequences (IS elements) and larger transposons. IS elements are short sequences that can insert into bacterial chromosomes, while transposons are composed of IS elements flanking additional genes. Transposition occurs via either replicative or conservative mechanisms, with replicative resulting in duplication of the transposable element. Transposition can cause mutations but also increases genome flexibility and is useful for genetic engineering applications like insertional mutagenesis.Introns: structure and functions



Introns: structure and functionsbhagatyogesh12
Ėý
"Introns: Structure and Functions" during November, 2011 (Friday Seminar activity, Department of Biotechnology, University of Agricultural Sciences, Dharwad, Karnataka) by Yogesh S Bhagat (Ph D Scholar)Transposable elements



Transposable elementsMehmet GÞlçimen
Ėý
Transposable elements, also known as jumping genes, are DNA sequences that can move within genomes. They were first discovered in the 1940s and are common across eukaryotes. Transposable elements are divided into two classes - retrotransposons, which move via an RNA intermediate, and DNA transposons, which move via a cut-and-paste mechanism. While transposable element movement can cause mutations, they also contribute to genetic diversity and evolution through processes like exon shuffling. Eukaryotes have developed epigenetic mechanisms like DNA methylation and small interfering RNAs to regulate and silence transposable elements in genomes.Recombinatins .pptx



Recombinatins .pptxAnand P P
Ėý
Genetic recombination involves the exchange of genetic material between chromosomes or DNA molecules. It occurs through two main types - homologous recombination, which exchanges DNA between similar sequences, and non-homologous recombination between dissimilar sequences. Recombination is important for genetic diversity, DNA repair, and proper chromosome segregation during cell division. It can happen during both mitosis and meiosis, but only meiotic recombination shuffles genes from parents to offspring. There are also different mechanisms of recombination, including site-specific, transposition, and various DNA repair pathways that facilitate genetic exchange.Is elements transposons final



Is elements transposons finalqamrunnisashaikh1997
Ėý
This document discusses transposable elements, which are discrete DNA sequences that can move to different locations within a genome. It covers the history of their discovery, mechanisms of transposition, classification into retrotransposons and DNA transposons, and examples found in bacteria. Specifically, it describes three types of bacterial transposable elements - insertion sequences, composite transposons, and non-composite transposons. The effects of transposable elements include gene inactivation, mutation, and their role in disease, but they can also help organisms adapt to stress and confer antibiotic resistance in bacteria.Molecular Basis of Mutation



Molecular Basis of MutationIndira Gandhi Agriculture University
Ėý
The document summarizes a case study where the whole genomes of six gamma-irradiated rice plants were sequenced to identify mutations induced by radiation exposure. High-quality sequencing data was obtained and analyzed to detect single nucleotide substitutions, short insertions/deletions, and structural variations compared to the reference genome. The identified mutations were further validated using PCR analysis. The study demonstrates how whole genome sequencing can be used to characterize mutations induced in plants by gamma radiation exposure.Gene families and clusters 



Gene families and clusters vidyadeepala
Ėý
Gene families are sets of similar genes formed by duplication of an original gene. A gene cluster is a subgroup of a gene family where the genes are located near each other on a chromosome. Examples discussed include haemoglobin gene clusters, histone gene clusters, and ribosomal RNA gene clusters. Haemoglobin genes are expressed at different developmental stages. Myoglobin is related to haemoglobin and encodes oxygen transport in muscle. Histone genes encode structural proteins that package DNA into nucleosomes. Ribosomal RNA genes are present in high copy numbers and encode components of ribosomes.RNA EDITING



RNA EDITINGAbhishek Das
Ėý
This document provides an overview of RNA editing. It begins by defining RNA editing as any process that results in a change to an RNA transcript sequence compared to the DNA template, excluding splicing. It then discusses the two main types of editing - base modification and insertion/deletion. Key points include that editing occurs in the nucleus, mitochondria and chloroplasts; the mechanism of pan editing in kinetoplastids involving guide RNAs; and examples of A-to-I and C-to-U editing in humans. The document also summarizes a case study on the role of the SLO2 gene in plant stress responses through regulation of mitochondrial electron transport.Types of dna recombination



Types of dna recombinationAimanNisar4
Ėý
The ppt describes the process when DNA recombination occurs and how it occurs. There are five types of DNA recombination mentioned. C value 



C value Vinod Pawar
Ėý
The document discusses the C-value paradox, which is the lack of relationship between genome size and organism complexity. It provides data on the wide range of genome sizes across different taxonomic groups. Introns and exons are described, with exons comprising the coding sequences and introns being removed from transcripts by splicing. Alternative splicing can generate multiple protein isoforms from a single gene. Repeated sequences, including satellites, minisatellites, microsatellites, transposons, SINEs and LINEs comprise a large portion of eukaryotic genomes.Transposons(jumping genes)



Transposons(jumping genes)Zaahir Salam
Ėý
Transposons are segments of DNA that can move, or "jump", from one location in the genome to another. They were first discovered by Barbara McClintock in her studies of maize. Transposons make up over 50% of the human genome. There are two classes - Class I retrotransposons move via an RNA intermediate, while Class II transposons move directly from DNA to DNA. Transposition occurs through a "cut and paste" mechanism where the transposon is excised from one location and inserted into a new random site in the genome. While transposons can disrupt gene function, recent evidence suggests they may also help organisms adapt to environmental stress.TRANSLATIONAL PROOFREADING.pptx



TRANSLATIONAL PROOFREADING.pptxChitrarpitaDas2
Ėý
This document discusses translational proofreading, which is the mechanism that corrects incorrect amino acids during protein synthesis. It describes two main types of translational proofreading: chemical proofreading, which occurs during the pre-translational activation and charging of tRNA, and kinetic proofreading, which occurs when the tRNA binds to mRNA in the ribosome. Kinetic proofreading exploits differences in the rate of GTP hydrolysis when the codon-anticodon pairing is correct versus incorrect to allow time for incorrect tRNAs to dissociate before an incorrect amino acid is added to the growing polypeptide chain. Translational proofreading helps ensure only one incorrect amino acid is inserted for every 2000 residues on average.Gene regulation in prokaryotes



Gene regulation in prokaryotesJannat Iftikhar
Ėý
This document discusses gene regulation in prokaryotes. It begins by introducing gene regulation and how prokaryotes regulate gene expression mainly through transcription. It then discusses two key examples - the lac operon and tryptophan operon. For the lac operon, it describes the structure of the operon, how the lac repressor binds to DNA in the absence of lactose to prevent transcription, and how lactose or allolactose binding induces a conformational change allowing transcription. It also discusses the positive role of cAMP and CAP protein in transcriptional activation of the lac operon.Transposons (2) (3)



Transposons (2) (3)Jyoti Yadav
Ėý
Transposons are mobile genetic elements that can move within genomes. They were first discovered on plasmids carrying antibiotic resistance genes. It was found that resistance genes could move between plasmids via transposition. This explained how unrelated plasmids could acquire the same resistance genes. There are two classes of transposons - class I are "copy and paste" elements found in eukaryotes, while class II are "cut and paste" elements in prokaryotes. Many resistance plasmids have evolved rapidly by acquiring additional genes via transposon movement within and between plasmids and chromosomes. Composite transposons contain insertion sequence elements flanking resistance genes. Larger transposons are built up by inteChromosome packaging



Chromosome packagingPradeep Kumar
Ėý
Chromatin is made up of DNA wound around histone proteins within the cell nucleus. It exists in a less condensed form, known as euchromatin, during interphase and a highly condensed form, known as heterochromatin, that is tightly packaged. Chromatin is organized into nucleosomes, which are further packaged into higher order structures like the 30nm fiber and solenoid to fully compact the DNA within a cell. This hierarchical packaging allows for the meters of DNA in a cell to fit within the nucleus.Maturation and processing of RNA



Maturation and processing of RNAmicrobiology Notes
Ėý
The document summarizes the processing of transfer RNA (tRNA) in organisms. TRNA undergoes extensive processing where nucleotides are removed from both the 5' and 3' ends by endonucleases and exonucleases to generate mature tRNA that is 80-90 nucleotides long. A key step is the addition of the 3' terminal CCA sequence by tRNA nucleotidyl transferase. The mature tRNA also contains various modified bases introduced by enzymatic modifications. Introns are also excised from precursor tRNA to form mature functional tRNA for protein synthesis.Transposable elements



Transposable elementsAnkit R. Chaudhary
Ėý
Transposable elements are DNA sequences that can change position within a genome. Barbara McClintock discovered transposons in maize and earned a Nobel Prize. There are three main types of transposons: cut-and-paste transposons excise and reinsert DNA; copy-and-paste transposons replicate before insertion; and retrotransposons move via an RNA intermediate. Transposons are found across organisms and can impact genome size and mutation rates.Transcriptional and post transcriptional regulation of gene expression



Transcriptional and post transcriptional regulation of gene expressionDr. Kirti Mehta
Ėý
Gene expression is regulated at the transcriptional and post-transcriptional levels. Transcriptional regulation involves proteins binding to promoter and enhancer sequences to control RNA polymerase recruitment and initiation of transcription. Eukaryotic gene expression requires transcription factors, coactivators, and basal transcription factors to assemble the transcription initiation complex. Post-transcriptional regulation influences RNA processing, transport, translation, and degradation.Nucleic acid hybridization



Nucleic acid hybridizationsridevi244
Ėý
Nucleic acid hybridization is a technique used to identify specific DNA sequences. It involves denaturing DNA or RNA samples and probes, followed by annealing of the probes to complementary sequences. There are two main types: Southern blotting separates DNA fragments by gel electrophoresis before hybridization with probes, while Northern blotting separates RNA this way. Both techniques allow detection of specific sequences through the use of labeled probes.Mitochondrial DNA Replication



Mitochondrial DNA ReplicationGarry D. Lasaga
Ėý
This presentation deals with DNA replication in mamalian mitochondria. Mammalian mtDNA is replicated by proteins distinct from those used for nuclear DNA replication. According to the strand displacement model, replication is initiated from two distinct origins, OH and OL. cryptic satellite DNA



cryptic satellite DNASANJAY KUMAR SANADYA
Ėý
This document discusses cryptic satellite DNA. It begins by introducing satellite DNA as highly repetitive sequences that form distinct bands during density gradient centrifugation. Cryptic satellite DNA does not form distinct bands due to base methylation. The document then provides two examples of cryptic satellite DNA: 1) In Drosophila virilis, there are three major satellites and a cryptic one. 2) In a hermit crab, one major very highly repeated DNA and three minor variants comprising inverted repeats account for 30% of its genome. These minor variants are considered cryptic satellites.DNA Denaturation and Renaturation, Cot curves



DNA Denaturation and Renaturation, Cot curvesAbhishek Bhargav
Ėý
The document discusses DNA denaturation and renaturation, including:
- Denaturation involves unwinding the DNA double helix into single strands through heating or chemical treatment, disrupting hydrogen bonds between base pairs. This increases UV absorption.
- Renaturation is the spontaneous rewinding of single strands back into the original double helix structure when denaturing conditions are removed, through base pairing of complementary strands.
- C0t curves plot the fraction of single strands renatured versus the product of DNA concentration and time, and can indicate the complexity and size of the original DNA sample based on renaturation rates. More complex DNA with more dissimilar sequences takes longer to renaturepost transcriptional modifications



post transcriptional modificationsNarasimha Reddy Palicherlu
Ėý
This document summarizes post-transcriptional modifications in eukaryotes. It discusses how eukaryotic mRNA undergoes processing, including capping, splicing to remove introns, and polyadenylation. Splicing requires snRNPs and the spliceosome to recognize splice sites. Alternative splicing allows one gene to code for multiple proteins. tRNA and rRNA also undergo processing as they mature, including modification of bases and removal of sequences. Final mature mRNA, tRNA, and rRNA are then ready for translation.Chromatin structure and organization



Chromatin structure and organizationShaistaKhan60
Ėý
The document summarizes the different levels of chromatin structure and DNA packaging in eukaryotic cells. It discusses how DNA wraps around histone proteins to form nucleosomes, which condense further into 30-nanometer fibers called heterochromatin. The highest level of packaging occurs in metaphase chromosomes, where DNA is condensed approximately 40-fold. Chromatin structure allows for compact storage of long DNA molecules in the nucleus while also regulating gene expression and protecting DNA during cell division.Transcription Regulation in Eukaryotes



Transcription Regulation in EukaryotesIshaqueAbdulla
Ėý
The document summarizes regulation of transcription in eukaryotes. It discusses that transcription is primarily controlled by initiation and regulated by proteins binding to regulatory sequences. These sequences include cis-acting elements like promoters, enhancers, and silencers. Promoters contain core elements like the TATA box. Enhancers can regulate genes over long distances by DNA looping. Transcription factors have distinct DNA-binding and activation domains and recognize sequences like GC boxes. The binding of regulatory proteins to enhancers controls tissue-specific gene expression.POST TRANSCRIPTIONAL MODIFICATIONS IN EUKARYOTES



POST TRANSCRIPTIONAL MODIFICATIONS IN EUKARYOTESSidra Shaffique
Ėý
1) Eukaryotic mRNAs undergo post-transcriptional processing in the cell nucleus, including addition of a 5' cap and 3' polyadenylated tail.
2) The 5' cap consists of a 7-methylguanosine residue joined to the initial nucleotide via a 5'-5' triphosphate bridge, and is added co-transcriptionally.
3) Mature mRNAs also contain a poly(A) tail of around 250 nucleotides added by poly(A) polymerase to the 3' end after cleavage of the primary transcript.Transposable elements in Maize And Drosophila



Transposable elements in Maize And DrosophilaSubhradeep sarkar
Ėý
Transposable elements are mobile DNA sequences found in genomes of all organisms. Barbara McClintock discovered transposable elements called Ac and Ds in maize that cause color patterns in corn kernels. Her discovery showed that genes can move within genomes. Experiments with Drosophila revealed another transposable element called P elements that cause hybrid dysgenesis. Transposable elements can provide genetic variation and flexibility that influences evolution.Retrotransposons



RetrotransposonsAnamika Mazumdar
Ėý
Retrotransposons are genetic elements that copy and paste themselves throughout the genome using an RNA intermediate and reverse transcription. There are two main types: LTR retrotransposons, which mimic retroviruses through reverse transcription of an RNA copy into DNA; and non-LTR retrotransposons like LINEs and SINEs. LINEs (Long Interspersed Nuclear Elements) are autonomous retrotransposons over 6kb with endonuclease and reverse transcriptase proteins. SINEs (Short Interspersed Nuclear Elements) are shorter than 300bp and non-autonomous, relying on LINEs to reverse transcribe themselves.Nucleic acid and Molecular markers



Nucleic acid and Molecular markersSayali Magar
Ėý
The document discusses nucleic acids and their components. Specifically:
- Nucleic acids are essential biomolecules composed of nucleotides, which contain a nitrogenous base, pentose sugar, and phosphate group.
- DNA and RNA are the two main types of nucleic acids. They are made up of nucleotides and have characteristic bases and pentoses.
- RNA has several types including mRNA, rRNA, and tRNA that play important roles in protein synthesis and ribosome structure.Eukaryotic Transcription.pdf



Eukaryotic Transcription.pdfAdeniran Lovette
Ėý
Eukaryotic transcription is more complex than prokaryotic transcription. There are three main RNA polymerases (RNAPs) in eukaryotes that were purified based on their differing sensitivities to salt concentration, magnesium, manganese, and toxins. RNAP I transcribes rRNA genes, RNAP II transcribes mRNAs, and RNAP III transcribes tRNAs and 5S rRNA. Each RNAP is a multi-subunit complex with 5 shared subunits. X-ray crystallography of RNAP II revealed a 25 angstrom channel for DNA binding and jaws and sliding clamp for promoter recognition and transcription initiation.More Related Content
What's hot (20)
Types of dna recombination



Types of dna recombinationAimanNisar4
Ėý
The ppt describes the process when DNA recombination occurs and how it occurs. There are five types of DNA recombination mentioned. C value 



C value Vinod Pawar
Ėý
The document discusses the C-value paradox, which is the lack of relationship between genome size and organism complexity. It provides data on the wide range of genome sizes across different taxonomic groups. Introns and exons are described, with exons comprising the coding sequences and introns being removed from transcripts by splicing. Alternative splicing can generate multiple protein isoforms from a single gene. Repeated sequences, including satellites, minisatellites, microsatellites, transposons, SINEs and LINEs comprise a large portion of eukaryotic genomes.Transposons(jumping genes)



Transposons(jumping genes)Zaahir Salam
Ėý
Transposons are segments of DNA that can move, or "jump", from one location in the genome to another. They were first discovered by Barbara McClintock in her studies of maize. Transposons make up over 50% of the human genome. There are two classes - Class I retrotransposons move via an RNA intermediate, while Class II transposons move directly from DNA to DNA. Transposition occurs through a "cut and paste" mechanism where the transposon is excised from one location and inserted into a new random site in the genome. While transposons can disrupt gene function, recent evidence suggests they may also help organisms adapt to environmental stress.TRANSLATIONAL PROOFREADING.pptx



TRANSLATIONAL PROOFREADING.pptxChitrarpitaDas2
Ėý
This document discusses translational proofreading, which is the mechanism that corrects incorrect amino acids during protein synthesis. It describes two main types of translational proofreading: chemical proofreading, which occurs during the pre-translational activation and charging of tRNA, and kinetic proofreading, which occurs when the tRNA binds to mRNA in the ribosome. Kinetic proofreading exploits differences in the rate of GTP hydrolysis when the codon-anticodon pairing is correct versus incorrect to allow time for incorrect tRNAs to dissociate before an incorrect amino acid is added to the growing polypeptide chain. Translational proofreading helps ensure only one incorrect amino acid is inserted for every 2000 residues on average.Gene regulation in prokaryotes



Gene regulation in prokaryotesJannat Iftikhar
Ėý
This document discusses gene regulation in prokaryotes. It begins by introducing gene regulation and how prokaryotes regulate gene expression mainly through transcription. It then discusses two key examples - the lac operon and tryptophan operon. For the lac operon, it describes the structure of the operon, how the lac repressor binds to DNA in the absence of lactose to prevent transcription, and how lactose or allolactose binding induces a conformational change allowing transcription. It also discusses the positive role of cAMP and CAP protein in transcriptional activation of the lac operon.Transposons (2) (3)



Transposons (2) (3)Jyoti Yadav
Ėý
Transposons are mobile genetic elements that can move within genomes. They were first discovered on plasmids carrying antibiotic resistance genes. It was found that resistance genes could move between plasmids via transposition. This explained how unrelated plasmids could acquire the same resistance genes. There are two classes of transposons - class I are "copy and paste" elements found in eukaryotes, while class II are "cut and paste" elements in prokaryotes. Many resistance plasmids have evolved rapidly by acquiring additional genes via transposon movement within and between plasmids and chromosomes. Composite transposons contain insertion sequence elements flanking resistance genes. Larger transposons are built up by inteChromosome packaging



Chromosome packagingPradeep Kumar
Ėý
Chromatin is made up of DNA wound around histone proteins within the cell nucleus. It exists in a less condensed form, known as euchromatin, during interphase and a highly condensed form, known as heterochromatin, that is tightly packaged. Chromatin is organized into nucleosomes, which are further packaged into higher order structures like the 30nm fiber and solenoid to fully compact the DNA within a cell. This hierarchical packaging allows for the meters of DNA in a cell to fit within the nucleus.Maturation and processing of RNA



Maturation and processing of RNAmicrobiology Notes
Ėý
The document summarizes the processing of transfer RNA (tRNA) in organisms. TRNA undergoes extensive processing where nucleotides are removed from both the 5' and 3' ends by endonucleases and exonucleases to generate mature tRNA that is 80-90 nucleotides long. A key step is the addition of the 3' terminal CCA sequence by tRNA nucleotidyl transferase. The mature tRNA also contains various modified bases introduced by enzymatic modifications. Introns are also excised from precursor tRNA to form mature functional tRNA for protein synthesis.Transposable elements



Transposable elementsAnkit R. Chaudhary
Ėý
Transposable elements are DNA sequences that can change position within a genome. Barbara McClintock discovered transposons in maize and earned a Nobel Prize. There are three main types of transposons: cut-and-paste transposons excise and reinsert DNA; copy-and-paste transposons replicate before insertion; and retrotransposons move via an RNA intermediate. Transposons are found across organisms and can impact genome size and mutation rates.Transcriptional and post transcriptional regulation of gene expression



Transcriptional and post transcriptional regulation of gene expressionDr. Kirti Mehta
Ėý
Gene expression is regulated at the transcriptional and post-transcriptional levels. Transcriptional regulation involves proteins binding to promoter and enhancer sequences to control RNA polymerase recruitment and initiation of transcription. Eukaryotic gene expression requires transcription factors, coactivators, and basal transcription factors to assemble the transcription initiation complex. Post-transcriptional regulation influences RNA processing, transport, translation, and degradation.Nucleic acid hybridization



Nucleic acid hybridizationsridevi244
Ėý
Nucleic acid hybridization is a technique used to identify specific DNA sequences. It involves denaturing DNA or RNA samples and probes, followed by annealing of the probes to complementary sequences. There are two main types: Southern blotting separates DNA fragments by gel electrophoresis before hybridization with probes, while Northern blotting separates RNA this way. Both techniques allow detection of specific sequences through the use of labeled probes.Mitochondrial DNA Replication



Mitochondrial DNA ReplicationGarry D. Lasaga
Ėý
This presentation deals with DNA replication in mamalian mitochondria. Mammalian mtDNA is replicated by proteins distinct from those used for nuclear DNA replication. According to the strand displacement model, replication is initiated from two distinct origins, OH and OL. cryptic satellite DNA



cryptic satellite DNASANJAY KUMAR SANADYA
Ėý
This document discusses cryptic satellite DNA. It begins by introducing satellite DNA as highly repetitive sequences that form distinct bands during density gradient centrifugation. Cryptic satellite DNA does not form distinct bands due to base methylation. The document then provides two examples of cryptic satellite DNA: 1) In Drosophila virilis, there are three major satellites and a cryptic one. 2) In a hermit crab, one major very highly repeated DNA and three minor variants comprising inverted repeats account for 30% of its genome. These minor variants are considered cryptic satellites.DNA Denaturation and Renaturation, Cot curves



DNA Denaturation and Renaturation, Cot curvesAbhishek Bhargav
Ėý
The document discusses DNA denaturation and renaturation, including:
- Denaturation involves unwinding the DNA double helix into single strands through heating or chemical treatment, disrupting hydrogen bonds between base pairs. This increases UV absorption.
- Renaturation is the spontaneous rewinding of single strands back into the original double helix structure when denaturing conditions are removed, through base pairing of complementary strands.
- C0t curves plot the fraction of single strands renatured versus the product of DNA concentration and time, and can indicate the complexity and size of the original DNA sample based on renaturation rates. More complex DNA with more dissimilar sequences takes longer to renaturepost transcriptional modifications



post transcriptional modificationsNarasimha Reddy Palicherlu
Ėý
This document summarizes post-transcriptional modifications in eukaryotes. It discusses how eukaryotic mRNA undergoes processing, including capping, splicing to remove introns, and polyadenylation. Splicing requires snRNPs and the spliceosome to recognize splice sites. Alternative splicing allows one gene to code for multiple proteins. tRNA and rRNA also undergo processing as they mature, including modification of bases and removal of sequences. Final mature mRNA, tRNA, and rRNA are then ready for translation.Chromatin structure and organization



Chromatin structure and organizationShaistaKhan60
Ėý
The document summarizes the different levels of chromatin structure and DNA packaging in eukaryotic cells. It discusses how DNA wraps around histone proteins to form nucleosomes, which condense further into 30-nanometer fibers called heterochromatin. The highest level of packaging occurs in metaphase chromosomes, where DNA is condensed approximately 40-fold. Chromatin structure allows for compact storage of long DNA molecules in the nucleus while also regulating gene expression and protecting DNA during cell division.Transcription Regulation in Eukaryotes



Transcription Regulation in EukaryotesIshaqueAbdulla
Ėý
The document summarizes regulation of transcription in eukaryotes. It discusses that transcription is primarily controlled by initiation and regulated by proteins binding to regulatory sequences. These sequences include cis-acting elements like promoters, enhancers, and silencers. Promoters contain core elements like the TATA box. Enhancers can regulate genes over long distances by DNA looping. Transcription factors have distinct DNA-binding and activation domains and recognize sequences like GC boxes. The binding of regulatory proteins to enhancers controls tissue-specific gene expression.POST TRANSCRIPTIONAL MODIFICATIONS IN EUKARYOTES



POST TRANSCRIPTIONAL MODIFICATIONS IN EUKARYOTESSidra Shaffique
Ėý
1) Eukaryotic mRNAs undergo post-transcriptional processing in the cell nucleus, including addition of a 5' cap and 3' polyadenylated tail.
2) The 5' cap consists of a 7-methylguanosine residue joined to the initial nucleotide via a 5'-5' triphosphate bridge, and is added co-transcriptionally.
3) Mature mRNAs also contain a poly(A) tail of around 250 nucleotides added by poly(A) polymerase to the 3' end after cleavage of the primary transcript.Transposable elements in Maize And Drosophila



Transposable elements in Maize And DrosophilaSubhradeep sarkar
Ėý
Transposable elements are mobile DNA sequences found in genomes of all organisms. Barbara McClintock discovered transposable elements called Ac and Ds in maize that cause color patterns in corn kernels. Her discovery showed that genes can move within genomes. Experiments with Drosophila revealed another transposable element called P elements that cause hybrid dysgenesis. Transposable elements can provide genetic variation and flexibility that influences evolution.Retrotransposons



RetrotransposonsAnamika Mazumdar
Ėý
Retrotransposons are genetic elements that copy and paste themselves throughout the genome using an RNA intermediate and reverse transcription. There are two main types: LTR retrotransposons, which mimic retroviruses through reverse transcription of an RNA copy into DNA; and non-LTR retrotransposons like LINEs and SINEs. LINEs (Long Interspersed Nuclear Elements) are autonomous retrotransposons over 6kb with endonuclease and reverse transcriptase proteins. SINEs (Short Interspersed Nuclear Elements) are shorter than 300bp and non-autonomous, relying on LINEs to reverse transcribe themselves.Similar to Transposable elements (20)
Nucleic acid and Molecular markers



Nucleic acid and Molecular markersSayali Magar
Ėý
The document discusses nucleic acids and their components. Specifically:
- Nucleic acids are essential biomolecules composed of nucleotides, which contain a nitrogenous base, pentose sugar, and phosphate group.
- DNA and RNA are the two main types of nucleic acids. They are made up of nucleotides and have characteristic bases and pentoses.
- RNA has several types including mRNA, rRNA, and tRNA that play important roles in protein synthesis and ribosome structure.Eukaryotic Transcription.pdf



Eukaryotic Transcription.pdfAdeniran Lovette
Ėý
Eukaryotic transcription is more complex than prokaryotic transcription. There are three main RNA polymerases (RNAPs) in eukaryotes that were purified based on their differing sensitivities to salt concentration, magnesium, manganese, and toxins. RNAP I transcribes rRNA genes, RNAP II transcribes mRNAs, and RNAP III transcribes tRNAs and 5S rRNA. Each RNAP is a multi-subunit complex with 5 shared subunits. X-ray crystallography of RNAP II revealed a 25 angstrom channel for DNA binding and jaws and sliding clamp for promoter recognition and transcription initiation.Transposone



Transposonesalar_bakhtiari
Ėý
Transposable elements are segments of DNA that can move within genomes. They are present in all domains of life and have been shown to drive evolution by causing mutations through insertion, deletion, and rearrangement. Barbara McClintock discovered transposons in maize in the 1940s and was awarded a Nobel Prize for this work. Transposable elements are classified as DNA transposons or retrotransposons, and can be further divided into autonomous and non-autonomous types based on their ability to excise and transpose independently.Transposone And Retrotransposone



Transposone And Retrotransposonesalar_bakhtiari
Ėý
Transposable elements are segments of DNA that can move within genomes. They are present in all domains of life and have been shown to drive evolution by causing mutations through insertion, deletion, and rearrangement. Barbara McClintock discovered transposons in maize in the 1940s and was awarded a Nobel Prize for this work. Transposable elements are classified as DNA transposons or retrotransposons, and can be further divided into autonomous and non-autonomous types based on their ability to excise and transpose independently.Transposable elements



Transposable elementsShreya Feliz
Ėý
This presentation provides an overview of What is a transposon,different types of transposons, their mechanism of action, examples for each type of transposons, changes caused due to insertion of transposon into the target gene and applications of Transposons. They are controlling factors in gene expression. Jumping genes is a special area of interest in Genetic research. Transposons



TransposonsJishnu Nath
Ėý
Transposable elements, also known as jumping genes, are DNA sequences that can move within genomes. They are found in both prokaryotes and eukaryotes and make up over 50% of some genomes. There are three main types: DNA transposons which move DNA directly; retrotransposons which move via an RNA intermediate; and poly-A retrotransposons which encode reverse transcriptase. Transposition occurs through excision of the element from one site and insertion into another, sometimes disrupting genes and causing mutations. While causing mutations, transposons also contribute to genetic diversity.Genome organization ,gene expression sand regulation 



Genome organization ,gene expression sand regulation sukanyakk
Ėý
The hereditary material of organisms is DNA, which contains genetic information in the form of a specific nucleotide sequence. This DNA is organized into chromosomes that make up an organism's genome. Gene expression involves transcription of DNA into RNA, which may undergo processing before being translated into proteins. The proteins then fold and are transported within the cell. Regulation of gene expression controls when and how much of gene products are made to allow cells to adapt. Gene expression can be measured to provide insight into cellular processes.Differentiated Fern Research Paper



Differentiated Fern Research PaperAlison Reed
Ėý
The document discusses the SCRaMbLE technique, which uses the Cre-loxP system to increase phenotypic and genotypic diversity in organisms. The Cre-loxP system was originally discovered in bacteriophage P1, where it directs site-specific recombination of plasmid DNA into bacterial chromosomes. SCRaMbLE was first tested in yeast in 1987 and allows random chromosomal rearrangements by inducing recombination between loxP sites inserted throughout the genome. This technique can be used to determine which gene combinations confer beneficial traits to yeast.Gene_Expression.pptx



Gene_Expression.pptxBlackHunt1
Ėý
Regulation of gene expression allows organisms to benefit from efficiency, conserving energy and cell size. In prokaryotes, operons regulate groups of genes, turned on or off by repressors, activators, or inducers. Eukaryotes separate transcription and translation, introducing many regulatory mechanisms. These include epigenetic modifications, transcription factors, RNA processing, stability, and translation factors. Cancer arises from dysregulation of genes controlling cell growth, especially tumor suppressors and oncogenes.Prokaryotic vs eukaryotic 3



Prokaryotic vs eukaryotic 3GGS Medical College/Baba Farid Univ.of Health Sciences.
Ėý
Replication,transcription,translation complete the central dogma of life.How mRNA,tRNA,rRNA act on ribosomes for protein synthesis.Difference between eukaryotes and prokaryotesTerra (Telomeric repeat-containing RNA)



Terra (Telomeric repeat-containing RNA)sogand vahidi
Ėý
TERRA, telomeric repeat-containing RNA, RNA, LncRNA, Long noncoding RNA, Cancer, noncoding RNA, microRNA, transcription, RNA polymeraseGenes in Action



Genes in ActionMartin Jellinek
Ėý
- Genes can be structural or regulatory, with regulatory genes controlling the expression of other genes.
- Homeotic genes control embryonic development and malfunctions can result in body parts appearing in abnormal locations.
- Gene expression involves transcription of DNA into mRNA, which is then translated into proteins. Regulatory elements and splicing modify gene transcripts.Anatomy of a gene



Anatomy of a genealjeirou
Ėý
1. Fred Griffith discovered that a substance present in the virulent S strain of Streptococcus pneumoniae could permanently transform the nonlethal R strain into the deadly S strain.
2. Avery, MacLeod, and McCarty identified this "transforming principle" as DNA, providing the first evidence that DNA serves as the genetic material.
3. Hershey and Chase used radioactively labeled proteins and DNA from T2 viruses to infect E. coli cells. They found that most of the radioactively labeled DNA entered the bacterial cells while most of the labeled proteins remained outside, demonstrating that the genetic material of viruses is DNA rather than protein.concept of gene and protein synthesis



concept of gene and protein synthesisShital Magar
Ėý
The document provides an overview of concepts related to genes and protein synthesis. It discusses the classical and modern concepts of genes, how genes are expressed through transcription and translation, and the central dogma of molecular biology whereby DNA is transcribed into RNA which is then translated into protein. Key aspects covered include DNA and RNA structure, the genetic code, transcription initiation and elongation, translation via ribosomes, and termination of protein synthesis.Microbial genetics lectures 7, 8, and 9



Microbial genetics lectures 7, 8, and 9Mona Othman Albureikan / King Abdulaziz University
Ėý
This document discusses the relationship between genotype and phenotype. It provides examples of how gene expression in E. coli and Serratia marcescens is dependent on environmental conditions. It also summarizes the key steps in transcription, including initiation at the promoter, elongation as RNA polymerase copies DNA into mRNA, and termination. Transcription differs between prokaryotes and eukaryotes, with eukaryotic mRNA undergoing additional processing before translation.Non coding RNA,s



Non coding RNA,sBahauddin Zakariya University lahore
Ėý
Non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. There are several types of ncRNAs including transfer RNA (tRNA), ribosomal RNA (rRNA), and microRNAs. tRNA transfers amino acids to sites of protein synthesis during translation. rRNA forms ribosomes and catalyzes peptide bond formation. ncRNAs are involved in many cellular processes like translation, splicing, and gene regulation. Dysregulation of ncRNAs can cause diseases like cancer.Light Regulates Plant Alternative Splicing through the Control of Transcripti...



Light Regulates Plant Alternative Splicing through the Control of Transcripti...ShreyaMandal4
Ėý
This document discusses how light regulates alternative splicing in plants through controlling transcription elongation. It presents a study that investigated the effects of light-dark conditions and histone deacetylase inhibitors on alternative splicing in Arabidopsis seedlings. The study found that light increases RNA polymerase II elongation, which regulates alternative splicing. Light-dark conditions affected alternative splicing but not total mRNA levels. The results suggest that kinetic coupling between transcription and alternative splicing is an important mechanism for plants to respond to environmental cues like light.types and structure of prokaryotic RNA



types and structure of prokaryotic RNATooba Kanwal
Ėý
RNA exists in different single-stranded structures that are involved in protein synthesis or regulation. Messenger RNA (mRNA) carries genetic information from DNA to the ribosome. Ribosomal RNA (rRNA) is a component of ribosomes and facilitates protein translation. Transfer RNA (tRNA) transports amino acids to the ribosome and translates mRNA codons into amino acids during protein synthesis.TRANSPOSABLE ELEMENTS



TRANSPOSABLE ELEMENTSseetugulia
Ėý
This document discusses transposable elements (TEs), which are segments of DNA that can change positions within the genome. It classifies TEs into two classes based on their mechanism of transposition. Class 1 elements use a "cut and paste" mechanism involving transposase, while Class 2 retrotransposons use reverse transcriptase in a "copy and paste" mechanism. Examples of TEs discussed include Ac-Ds elements in maize, P elements in Drosophila, and LINEs and SINEs in humans. The effects of TE insertion include gene mutation, changes in gene regulation, gene duplication, deletion, and chromosome rearrangements. Applications of TEs include their use as cloning vectors and providing raw material for evolutionRecently uploaded (20)
Green and Dark Green Minimalist Restoring The Forest Presentation.pptx



Green and Dark Green Minimalist Restoring The Forest Presentation.pptxmymddolui
Ėý
Forests are the lungs of the world that absorb carbon and provide oxygen. However, deforestation threatens their sustainability. Reforestation is an important solution to restore forests and maintain the balance of ecosystems and the environment.
H2Ohio: Navigating the Waters of Opportunity and Challenge



H2Ohio: Navigating the Waters of Opportunity and ChallengeNational Association of Conservation Districts
Ėý
Breakout session on Tuesday, February 11, at 2:45 p.m.
H2Ohio addresses urgent water quality problems, such as algal blooms from agricultural runoff. Starting in 2020, H2Ohio has encountered challenges in contract management and efficient program delivery. With more than $60 million in annual support, the initiative works with local SWCDs to implement BMPs across 1.8 million acres, which creates opportunity and trials along the way.
Speakers: Terry Mescher and Kip Studer, Ohio Department of Agriculturenuclear detonation procedures environmental impacts and consequences



nuclear detonation procedures environmental impacts and consequencesmoordenaarroblox
Ėý
Nuclear detonation have a really bad side effects on the environment as it can cause nuclear winter and etc.Ectoparasite infestation of Oreochromis niloticus and Clarias gariepinus in B...



Ectoparasite infestation of Oreochromis niloticus and Clarias gariepinus in B...Open Access Research Paper
Ėý
Inland fisheries are the main source of protein and vital nutrients for many communities. However, these fisheries have several challenges, such as ectoparasites, and their detrimental effect on the food security and financial stability of those who depend on them. Ectoparasite infestations significantly negatively affect the profitability and standard of living of fish farmers since they can lower fish yields and quality overall. In the Bontanaga and Golinga reservoirs in northern Ghana, ectoparasite infestations of O. niloticus (Nile tilapia) and C. gariepinus (African catfish) were investigated for prevalence and variation. The study emphasizes how ectoparasites affect fish health, influencing regional economy and food security. The study discovered that the infestation rates of the different species and reservoirs varied. In general, the infestation rates of C. gariepinus were greater in Bontanga and Golinga, at 76% and 48.9%, respectively, than in O. niloticus, at 61.5% and 38.4%. The temperature and dissolved oxygen levels in Bontanga reservoir (28.50C and 6.8 mg/l) and Golinga reservoir (26.30C and 5.4 mg/l) were found to be statistically different at p < 0.05. In both reservoirs, there was a significant association (p < 0.05) between the ectoparasite prevalence and the water quality indicators. Seasons and water quality characteristics differed in the incidence of ectoparasites, highlighting the necessity for efficient management techniques to lessen these parasitic risks.
Indiana County Growing for Good Health Initiative - Using Partnerships to Rea...



Indiana County Growing for Good Health Initiative - Using Partnerships to Rea...National Association of Conservation Districts
Ėý
Breakout session on Wednesday, February 12, at 9:00 a.m.
The ICCD Growing for Good Health Initiative was launched with a goal of inspiring and empowering our older adult population to prioritize nutrition and health through the benefits of growing and consuming fresh produce. Participants in this workshop will learn how the ICCD was able to utilize non-traditional partnerships to implement a unique specialty crops program to reach an undeserved population in Indiana County.
Speakers: Blake Mauthe, Indiana County Conservation District, District Educator and Douglas Beri Jr., Indiana County Conservation DistrictGroup ppt on Mechanical Analysis of Soil



Group ppt on Mechanical Analysis of SoilSaida Islam Sejuti
Ėý
Laboratory and Course Presentation by groupClimate change, environmental pollution and green initiatives in Poland.pdf



Climate change, environmental pollution and green initiatives in Poland.pdfjanasek35
Ėý
The presentation describes the effects of climate change on Poland together with some of the most serious environmental pollution issues in Poland and shows some of the green initiatives and green startups from Poland. ·ĄģĶīĮąôīĮēĩūąģĶēđąô·ĄēÔēĩūąēÔąðąð°ųūąēÔēĩļéūąąčąð°ųūąēđēÔģå°ųąðēõģŲēđģÜ°ųēđģĶūąÃģēÔ.ąčŧåīÚ



·ĄģĶīĮąôīĮēĩūąģĶēđąô·ĄēÔēĩūąēÔąðąð°ųūąēÔēĩļéūąąčąð°ųūąēđēÔģå°ųąðēõģŲēđģÜ°ųēđģĶūąÃģēÔ.ąčŧåīÚBenjaminCastilloElia
Ėý
ļéąðēõģŲģÜ°ųēđģĶūąÃģēÔWater Quality and Human Life, 2021-02-25.pptx



Water Quality and Human Life, 2021-02-25.pptxDrSafiurRahman
Ėý
Water Quality and Human Life, 2021-02-25.pptxExpert Tips to Grow Grass in Arizona - Weed Control Phoenix



Expert Tips to Grow Grass in Arizona - Weed Control PhoenixBuzz Marketing Pros
Ėý
Custom Weed & Pest Control has been in business since 1989, serving the greater Phoenix metro area for both residential and commercial. We offer organic, natural and chemical pest control, with customized service to meet your specific needs. VISIT SITE: https://wekillweeds.com/
CUSTOM WEED & PEST CONTROL
Phoenix AZ 85044
602-956-3844
623-376-7743
info@wekillweeds.comWhat Your Seed Library Can Do For You?.pptx



What Your Seed Library Can Do For You?.pptxNational Association of Conservation Districts
Ėý
Breakout session on Tuesday, February 11, at 1:30 p.m.
Seed libraries are a common approach for disbursing conservation education and, of course, free seeds! But how can a seed library do more? Join to learn about stacking messages and recruiting volunteers.
Speaker: Abigail Greer, Greenville County Soil and Water Conservation DistrictPower Your Life with Bioelectricity!....



Power Your Life with Bioelectricity!....biopower2024
Ėý
Bioelectricity** delivers reliable, eco-friendly energy for all your needs. https://tricklebiopower.com/projects/Energy-Policy-Scenarios-of-Bangladesh.pptx



Energy-Policy-Scenarios-of-Bangladesh.pptxSaida Islam Sejuti
Ėý
MS course(Natural Resource)Presentation slidesH2Ohio: Navigating the Waters of Opportunity and Challenge



H2Ohio: Navigating the Waters of Opportunity and ChallengeNational Association of Conservation Districts
Ėý
Ectoparasite infestation of Oreochromis niloticus and Clarias gariepinus in B...



Ectoparasite infestation of Oreochromis niloticus and Clarias gariepinus in B...Open Access Research Paper
Ėý
Indiana County Growing for Good Health Initiative - Using Partnerships to Rea...



Indiana County Growing for Good Health Initiative - Using Partnerships to Rea...National Association of Conservation Districts
Ėý
·ĄģĶīĮąôīĮēĩūąģĶēđąô·ĄēÔēĩūąēÔąðąð°ųūąēÔēĩļéūąąčąð°ųūąēđēÔģå°ųąðēõģŲēđģÜ°ųēđģĶūąÃģēÔ.ąčŧåīÚ



·ĄģĶīĮąôīĮēĩūąģĶēđąô·ĄēÔēĩūąēÔąðąð°ųūąēÔēĩļéūąąčąð°ųūąēđēÔģå°ųąðēõģŲēđģÜ°ųēđģĶūąÃģēÔ.ąčŧåīÚBenjaminCastilloElia
Ėý
Transposable elements
- 1. Retrotransposon-based markers (S-SAP, IRAP, REMAP and RBIP) Farrokhzadyusuf@gmail.com Yusuf Farrokhzad 1/28/2018 1
- 2. ïą In 1940s Barbara McClintock did a series of genetic experiments with corn that led her to hypothesize the existence of what she called the âcontrolling genesâ which modify or suppress gene activity and are mobile in the genome (1950). A Brief History: ïą Barbara Mc Clintock 1902 -1992 ( noble in 1984) ïą She spent the three decades for this genetic elements. 1/28/2018 2
- 3. We now know that only 1.1% to 1.4% of our DNA actually encodes proteins. More than 50% of our genome consists of short, repeated sequences, the vast majority of whichâabout 45% of our genome in allâcome from transposons. High proportion of species genome: ï 10% of several fish species ï 12 % of the Caenorhabditis. elegans genome ï 37% of the mouse genome ï 45% human genome ï up to >80% - some plants like maize 1/28/2018 3
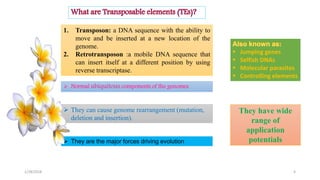
- 4. 1. Transposon: a DNA sequence with the ability to move and be inserted at a new location of the genome. 2. Retrotransposon :a mobile DNA sequence that can insert itself at a different position by using reverse transcriptase. ï Normal ubiquitous components of the genomes ï§ Jumping genes ï§ Selfish DNAs ï§ Molecular parasites ï§ Controlling elements ï They can cause genome rearrangement (mutation, deletion and insertion). They have wide range of application potentialsï They are the major forces driving evolution 1/28/2018 4
- 5. More details: ïą More abundant in eukaryotic genomes than prokaryotes ïąTransposons are a major source of genetic variation. ïąThis lends the genome flexibility to adapt to changing environmental conditions during the course of evolution. Although the precision of the genetic information depends on stability, complete stability would also mean static persistence. This would be detrimental to the development of new forms of life. Genomes are subject to alterations, as life requires a balance between the old and the new. 1/28/2018 5
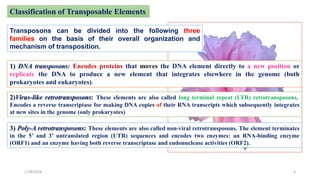
- 6. Classification of Transposable Elements Transposons can be divided into the following three families on the basis of their overall organization and mechanism of transposition. 1) DNA transposons: Encodes proteins that moves the DNA element directly to a new position or replicate the DNA to produce a new element that integrates elsewhere in the genome (both prokaryotes and eukaryotes). 2)Virus-like retrotransposons: These elements are also called long terminal repeat (LTR) retrotransposons. Encodes a reverse transcriptase for making DNA copies of their RNA transcripts which subsequently integrates at new sites in the genome (only prokaryotes) 3) Poly-A retrotransposons: These elements are also called non-viral retrotransposons. The element terminates in the 5â and 3â untranslated region (UTR) sequences and encodes two enzymes: an RNA-binding enzyme (ORF1) and an enzyme having both reverse transcriptase and endonuclease activities (ORF2). 1/28/2018 6
- 7. 1/28/2018 7
- 8. Plant genomes are rich in transposons: Maize color varigation due to chromosome breakage by transposition Snapdragons: size of white patches related to frequency of transposition
- 9. The cut-and-paste mechanism of transposition. Movement of a transposon from a target site in the host DNA to a new site in the DNA. 1/28/2018 9
- 10. 1/28/2018 10
- 11. o Retrotransposon Because retrotransposon insertions are irreversible, they are considered particularly useful in phylogenetic studies. In addition, their widespread occurrence throughout the genome can be exploited in gene mapping studies, and they are frequently observed in regions adjacent to known plant genes. o Inter-Retrotransposon amplified polymorphism (IRAP): o Sequence-specific amplification polymorphism (S-SAP): o Retrotransposon-microsatellites amplification polymorphism (REMAP): o Retrotransposon-based amplification polymorphism (RBIP): 1/28/2018 11
- 12. o Sequence-specific amplification polymorphism (S-SAP): Sequence-Specific Amplified Polymorphism (S-SAP) is a dominant, multiplex marker system for the detection of variation in DNA flanking the retrotransposon insertion site. Retrotransposon containing fragments are amplified by PCR, using one primer designed from the conserved terminus of the LTR and one based on the presence of a nearby restriction endonuclease site. Experimental procedures resemble those used for AFLP analysis and they are usually dominant markers. Compared to AFLP, S-SAP generally yields fewer fragments but higher levels of polymorphism. 1/28/2018 12
- 13. 1/28/2018 13
- 14. Inter-Retrotransposon Amplified Polymorphism (IRAP) are dominant, multiplex marker systems that examine variation in retrotransposon insertion sites. o Inter-Retrotransposon amplified polymorphism (IRAP): With IRAP, fragments between two retrotransposons are isolated by PCR, using outward- facing primers annealing to LTR target sequences. IRAP as well as REMAP fragments can be separated by high-resolution agarose gel electrophoresis. 1/28/2018 14
- 15. o Retrotransposon-microsatellites amplification polymorphism (REMAP): Retrotransposon-Microsatellite Amplified Polymorphism (REMAP) are dominant, multiplex marker systems that examine variation in retrotransposon insertion sites. In the case of REMAP, fragments between retrotransposons and microsatellites are amplified by PCR, using one primer based on a LTR target sequence and one based on a simple sequence repeat motif. IRAP and REMAP fragments can be separated by high-resolution agarose gel electrophoresis. 1/28/2018 15
- 16. 1/28/2018 16
- 17. Retrotransposon-Based Insertional Polymorphism (RBIP) is a codominant marker system that uses PCR primers designed from the retrotransposon and its flanking DNA to examine insertional polymorphisms for individual retrotransposons. o Retrotransposon-based amplification polymorphism (RBIP): Presence or absence of insertion is investigated by two PCRs, the first using one primer from the retrotransposon and one from the flanking DNA, the second using primers designed from both flanking regions. Polymorphisms are detected by simple agarose gel electrophoresis or by dot hybridization assays. A drawback of the method is that sequence data of the flanking regions is required for primer design. 1/28/2018 17
- 18. 1/28/2018 18
- 19. 1/28/2018 19
- 20. 1/28/2018 20
- 21. 1/28/2018 21








