Visualization of RNA 2D Structures - Structure/function relationships of H/ACA motifs: Analysis & Prediction
- 1. Visualization of RNA 2D Structures Structure-Function Relationships of H/ACA Motifs Fabrice Leclerc & Yann Ponty
- 2. 2D models (experimental: footprinting, fluorescence, SHAPE, etc; theoretical: MFold, RNAfold, etc) 2D representations from RNA 3D structures media to display: structural, functional, phylogenetic informations, etc RNAplot jViz.RNA
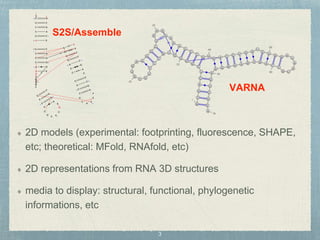
- 3. 2D models (experimental: footprinting, fluorescence, SHAPE, etc; theoretical: MFold, RNAfold, etc) 2D representations from RNA 3D structures media to display: structural, functional, phylogenetic informations, etc S2S/Assemble VARNA
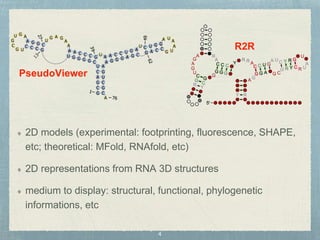
- 4. 2D models (experimental: footprinting, fluorescence, SHAPE, etc; theoretical: MFold, RNAfold, etc) 2D representations from RNA 3D structures medium to display: structural, functional, phylogenetic informations, etc PseudoViewer R2R
- 6. C: Stockholm (RFAM) D: Vienna (RNAfold) E: Pseudobase A: FASTA (aligned) B: CLUSTAL F: BPSEQ G: CONNECT (CT, CT2)
- 7. conformation 1 (ON) conformation 2 (OFF) VARNA Arc RIBOSWITCH graph
- 8. mutations, chemical probing data Aalberts & Jannen, RNA, 2013 base pairing 1 RNAbows
- 10. conservation VARNA*R2R * post-processingPerreault et al., PLoS One, 2011 Leclerc, Molecules, 2010 2D representation 2D representation (3D)
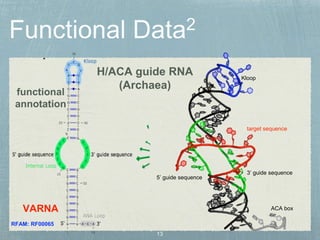
- 12. 3Ī» guide sequence 5Ī» guide sequence Kloop ACA box functional annotation H/ACA guide RNA (Archaea) VARNA RFAM: RF00065
- 13. 3Ī» guide sequence 5Ī» guide sequence Kloop ACA box target sequence functional annotation H/ACA guide RNA (Archaea) VARNA RFAM: RF00065
- 14. L7Ae Nop10 Cbf5 H/ACA guide RNA (Archaea) VARNA RFAM: RF00065 functional annotation
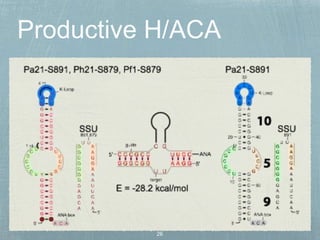
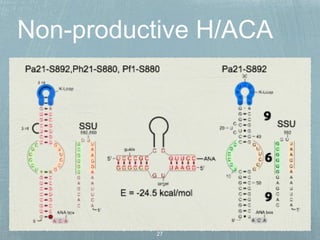
- 20. 7 H/ACA genes 11 H/ACA motifs Muller et al., NAR, 2008 Pyrococcus & Thermococcus
- 21. Muller et al., NAR, 2008
- 22. Muller et al., NAR, 2008
- 23. R2R
- 24. + -
- 27. standard and customized representations (edition, annotation) time saving for the generation of multiple representations time saving for the update of consensus structures qualitatif (quantitatif) tools to evaluate structure-function models
- 28. RFAM (VARNA), Comparative RNA Web Site & Project (RNA2DMap, CT, BPSEQ), The RNA Mapping Database - RMDB (RDAT, RNAstructure), ... packages: Vienna Package (RNAfold, RNAalifold, etc), Boulder Alignment Editor (VARNA), SAVor (RNAfold, RNAplot), S2S/Assemble 2D & 3D, Jalview 2D & 3D (VARNA, Jmol), ... recent tools: long RNAs: RNAfdl, pairing probabilities (alternative conformations): RNAbow, RNAllViewer, ...
- 29. Institut de G©”n©”tique & Microbiologie, Orsay Palaiseau