Barcelona sabatica
- 1. Protein loop classification using Artificial Neural Networks Armando Vieira1 and Baldomero Oliva2 1 ISEP and Centro de Física Computacional, Coimbra, Portugal www.defi.isep.ipp.pt/~asv 2 Structural Bioinformatics Laboratory (GRIB) IMIM/Universitat Pompeu Fabra, Barcelona, Spain
- 2. XXI: the century of BIO
- 3. BIOINFORMATICS joining two worlds apart
- 4. Outline Brief review of protein structure Statement of problem and why is so hard Data pre-processing, corrections, updates and beyond multiple alignments Neural Networks in protein structure prediction HLVQ Results and future work
- 5. Proteins All proteins are chains of 20 amino acids Not all chains of amino acids are proteins Fold rapidly and repeatedly Proteins are the machinery of live Essential to all (known) organisms
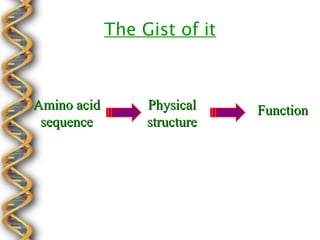
- 6. The Gist of it Amino acid Physical Function sequence structure
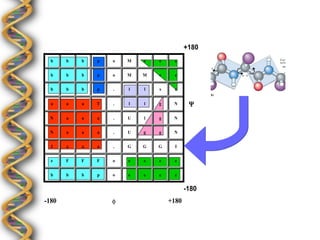
- 9. +180 b b b p o M e e e b b b p o M M e e b b b p . l l s e a a a T . l l g N ╬¿ N a a a . U l g N N a a a . U g g N I a a a . G G G I e F F F o e e e e b b b p o e e e e -180 -180 ¤å +180
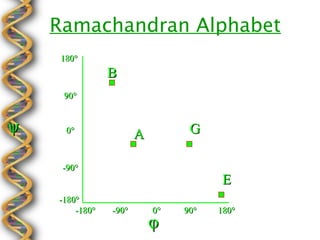
- 10. Ramachandran Alphabet 180┬░ B 90┬░ ¤ê 0┬░ A G -90┬░ E -180┬░ -180┬░ -90┬░ 0┬░ 90┬░ 180┬░ ¤å
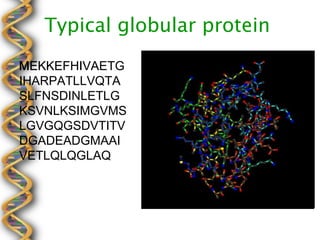
- 11. 5-letter alphabet Residue Sequence 3┬░ Structure MEKKEFHIVAET ACCDECBAABDE GIHARPATLLVQT CBDABCDBEABD ASLFNSDINLETL BCBDBAEBDBDB GKSVNLKSIMGV AEBABDCBBDBA MSLGVGQGSDVT DDCBDBCBDBEB ITVDGADEADGM DBCBBDCAABDE AAIVETLQLQGLA DCDCEAABACAA Q... AADCÔǪ
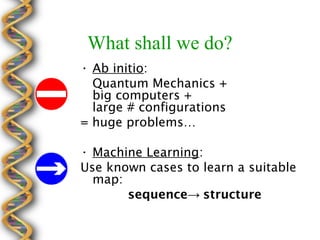
- 12. What shall we do?  Ab initio: Quantum Mechanics + big computers + large # configurations = huge problems  Machine Learning: Use known cases to learn a suitable map: sequence structure
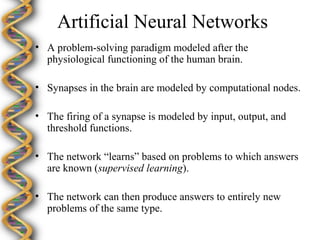
- 14. Artificial Neural Networks ÔÇó A problem-solving paradigm modeled after the physiological functioning of the human brain. ÔÇó Synapses in the brain are modeled by computational nodes. ÔÇó The firing of a synapse is modeled by input, output, and threshold functions. ÔÇó The network ÔÇ£learnsÔÇØ based on problems to which answers are known (supervised learning). ÔÇó The network can then produce answers to entirely new problems of the same type.
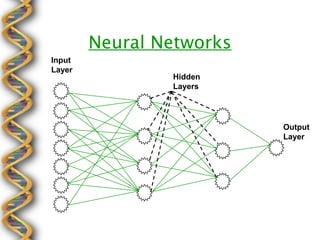
- 15. Neural Networks Input Layer Hidden Layers Output Layer
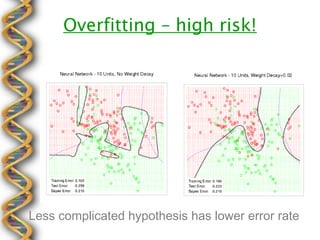
- 16. Overfitting ÔÇô high risk! Less complicated hypothesis has lower error rate
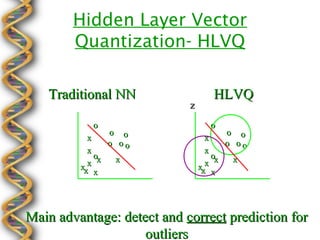
- 17. Hidden Layer Vector Quantization- HLVQ Traditional NN HLVQ z o o o o o o x x o oo o oo x x ox x ox x xxx xxx x x Main advantage: detect and correct prediction for outliers
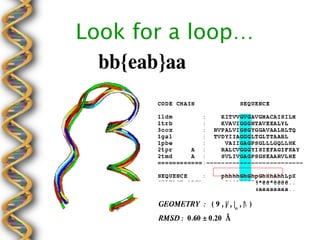
- 19. Look for a ▒¶┤Ã┤Ã▒ÞÔǪ
- 20. Geometry of the Motif
- 21. Loop Types ╬▒ÔêÆ╬▒ : ╬▒ -helix - ╬▒ -helix ╬▒ÔêÆ╬▓ : ╬▒ -helix ÔÇô ╬▓ strand ╬▓ÔêÆ╬▒ : ╬▓ strand - ╬▒ -helix ╬▓ -hairpin: ╬▓ strand - ╬▓ strand ╬▓ - link: ╬▓ strand - ╬▓ strand
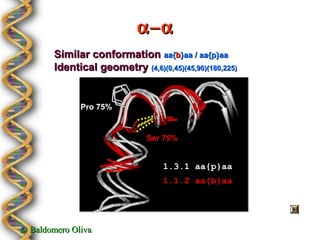
- 22. ╬▒ÔêÆ╬▒ Similar conformation aa{b}aa / aa{p}aa Identical geometry (4,6)(0,45)(45,90)(180,225) Pro 75% Ser 75% 1.3.1 aa{p}aa 1.1.2 aa{b}aa ┬® Baldomero Oliva
- 23. Class ╬▒ÔêÆ╬▒
- 24. ArchDB database ~ 20 000 loops classified into ~ 3000 classes. EE-3.4.1 Loop type - loop size . consensus . motif TASK: classify a loop from sequence alone If not possible, get as much information as possible
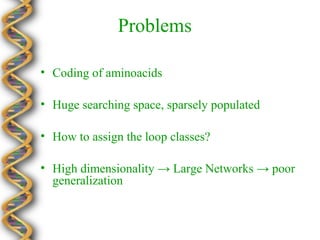
- 25. Problems  Coding of aminoacids  Huge searching space, sparsely populated  How to assign the loop classes?  High dimensionality  Large Networks  poor generalization
- 26. Aminoacid coding the classical way A  (1, 0, 0) C  (0, 1, 0) Y  (0, 0, 1) Useful but not efficient!!! I am working to improve it
- 27. Theory; but how about applications?!
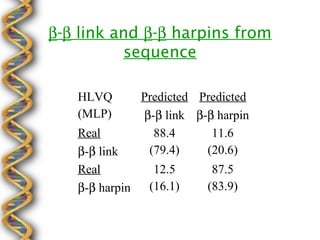
- 28. ╬▓-╬▓ link and ╬▓-╬▓ harpins from sequence HLVQ Predicted Predicted (MLP) ╬▓-╬▓ link ╬▓-╬▓ harpin Real 88.4 11.6 ╬▓-╬▓ link (79.4) (20.6) Real 12.5 87.5 ╬▓-╬▓ harpin (16.1) (83.9)
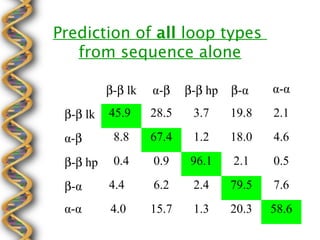
- 29. Prediction of all loop types from sequence alone ╬▓-╬▓ lk ╬▒-╬▓ ╬▓-╬▓ hp ╬▓-╬▒ ╬▒-╬▒ ╬▓-╬▓ lk 45.9 28.5 3.7 19.8 2.1 ╬▒-╬▓ 8.8 67.4 1.2 18.0 4.6 ╬▓-╬▓ hp 0.4 0.9 96.1 2.1 0.5 ╬▓-╬▒ 4.4 6.2 2.4 79.5 7.6 ╬▒-╬▒ 4.0 15.7 1.3 20.3 58.6
- 30. WhatÔÇÖs it all mean? Given a loop residue sequence, we can (usually) identify its native structure. Not ab initio: We cannot tell the structure of a novel sequence. HLVQ is superior to MLP
- 31. Future Work Better coding of aminoacids Larger sequences / low complexity Going beyond structure Clever alphabet that explore similarities Multiobjective Genetic Algorithms
- 32. Beyond Multiple Alignments  Alligments are good  but expensive and boring ...  Information contained in a multiple alignment can, in principle, be expressed using an adequate aminoacid coding scheme Sensibility  How? Genetic Algorithm
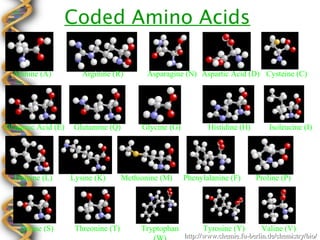
- 33. Coded Amino Acids Alanine (A) Arginine (R) Asparagine (N) Aspartic Acid (D) Cysteine (C) Glutamic Acid (E) Glutamine (Q) Glycine (G) Histidine (H) Isoleucine (I) Leucine (L) Lysine (K) Methionine (M) Phenylalanine (F) Proline (P) Serine (S) Threonine (T) Tryptophan Tyrosine (Y) Valine (V) http://www.chemie.fu-berlin.de/chemistry/bio/
- 34. ArchDB database Protein Data Bank (PDB) http://www.rcsb.org contains ~ 25 000 proteins with known structure of ~ 106 entries in SWISS-PROT ArchDB ~ 20 000 classified loops