Sequence Alignment - Data Bioinformatics Introduction
Download as PPTX, PDF0 likes21 views
Sequence Alignment
1 of 12
Download to read offline



Recommended
20100515 bioinformatics kapushesky_lecture07



20100515 bioinformatics kapushesky_lecture07Computer Science Club
Ěý
The document discusses pairwise sequence alignment methods. It defines key concepts like homology and orthology. It explains that dynamic programming is used to find optimal alignments through building a score matrix and backtracking. Global alignment finds the best match over full sequences while local alignment identifies regions of local similarity. Scoring systems like PAM matrices assign values based on substitutions and penalties for gaps.Seq alignment 



Seq alignment Nagendrasahu6
Ěý
Sequence alignment involves arranging biological sequences like DNA, RNA, or proteins to identify similar regions that may indicate functional, structural, or evolutionary relationships. There are two main types of sequence alignment: local alignment, which finds short, locally similar regions; and global alignment, which tries to match the full sequences. Sequence alignment is performed using algorithms like Needleman-Wunsch for global alignment and Smith-Waterman for local alignment. It can provide information about sequence homology and evolutionary relationships between sequences.Introduction to sequence alignment



Introduction to sequence alignmentKubuldinho
Ěý
An introductory lecture on sequence alignments.
Ideal for newbies, and also gurus of bioinformatics.
Feedback will be MUCH appreciated.Bioinformatics t4-alignments wim_vancriekingev2013



Bioinformatics t4-alignments wim_vancriekingev2013Prof. Wim Van Criekinge
Ěý
The document discusses dot plots and their use in bioinformatics. It explains that dot plots are a graphical representation that uses two sequences as axes and plots dots where regions of similarity are found based on a given threshold and window size. Dot plots can be used to visualize all similarities and repeats within and between sequences. Reducing window size and increasing stringency can reduce noise in dot plots. Available programs for generating dot plots are also mentioned.Bioinformatica t4-alignments



Bioinformatica t4-alignmentsProf. Wim Van Criekinge
Ěý
This document discusses dot plots and their use in bioinformatics. It begins by defining dot plots as a graphical representation that uses two sequences on orthogonal axes and plots dots where regions of similarity meet a given threshold within a window. Dot plots allow visualization of all structures in common between sequences or repeated/inverted structures within a sequence. The document provides an example dot plot creation script in Perl and discusses how to reduce noise in dot plots by increasing the window size or stringency. It notes common uses of dot plots like comparing genomic and cDNA sequences to predict exons. Finally, it provides some rules of thumb for effective dot plot analysis and lists available dot plot programs.sequence alignment



sequence alignmentammar kareem
Ěý
The document provides an overview of computational methods for sequence alignment. It discusses different types of sequence alignment including global and local alignment. It also describes various methods for sequence alignment, such as dot matrix analysis, dynamic programming algorithms (e.g. Needleman-Wunsch, Smith-Waterman), and word/k-tuple methods. Scoring matrices like PAM and BLOSUM that are used for sequence alignments are also explained.Gemoda



GemodaKyle Jensen
Ěý
The document describes the Gemoda algorithm for discovering motifs (patterns) in biomolecular data sequences. Gemoda is designed to be exhaustive in finding all maximal motifs and have descriptive power by using a generic, context-dependent definition of similarity. It proceeds in three steps: comparison of all pairwise windows to create a similarity graph, clustering similar windows into elementary motifs, and convolving the motifs to find longer, maximal motifs. Gemoda can be applied to problems like discovering protein domains, solving motif discovery challenges, and finding conserved structures in protein structures.Biomolecular Assignment Help



Biomolecular Assignment HelpNursing Assignment Help
Ěý
I am Mercy Knowles. Currently associated with nursingassignmenthelp.com as nursing homework helper. After completing my master's from Albany State University, USA, I was in search for an opportunity that expands my area of knowledge hence I decided to help students with their assignments. I have written several Biomolecular assignments till date to help students overcome numerous difficulties they face.An Efficient Biological Sequence Compression Technique Using LUT and Repeat ...



An Efficient Biological Sequence Compression Technique Using LUT and Repeat ...IOSR Journals
Ěý
This document presents two improved biological sequence compression algorithms that utilize a lookup table (LUT) and identification of tandem repeats in sequences. The first algorithm maps all possible 3-character combinations to ASCII characters using a 125-entry LUT. The second maps all possible 4-character combinations to ASCII characters using a 256-entry LUT. These algorithms aim to achieve high compression factors, saving percentages, and faster compression/decompression times compared to previous biological sequence compression methods.DNA Compression (Encoded using Huffman Encoding Method)



DNA Compression (Encoded using Huffman Encoding Method)Marwa Al-Rikaby
Ěý
The document discusses DNA compression algorithms. It describes the common components of most DNA compression algorithms, which include finding repeat segments in a DNA sequence, considering approximate repeats allowing for operations like substitutions, and selecting the best set of compatible repeats. It then provides an example demonstrating how these steps may be applied to a sample DNA sequence to identify repeat segments and encode them for compression.lecture4.ppt Sequence Alignmentaldf sdfsadf



lecture4.ppt Sequence Alignmentaldf sdfsadfalizain9604
Ěý
This document summarizes key concepts in sequence alignment including:
1) Sequence alignment involves finding the linear correspondence between symbols in one sequence to another that maximizes similarity. Dynamic programming is commonly used to compute optimal alignments.
2) BLAST is an extremely fast database search tool that uses heuristics like word matching to find local alignments and statistical analysis to assess significance.
3) Multiple sequence alignments make conserved features more apparent but are more difficult to compute than pairwise alignments. Progressive alignment gradually merges pairwise alignments based on a phylogenetic tree.Basics of bioinformatics



Basics of bioinformaticsAbhishek Vatsa
Ěý
Bioinformatics emerged from the marriage of computer science and molecular biology to analyze massive amounts of biological data, like that produced by the Human Genome Project. It uses algorithms and techniques from computer science to solve problems in molecular biology, like comparing genomic sequences to understand evolution. As genomic data exploded publicly, bioinformatics was needed to efficiently store, analyze, and make sense of this information, which has applications in molecular medicine, drug development, agriculture, and more.protein structure prediction in bioinformatics.ppt



protein structure prediction in bioinformatics.pptDrSudha2
Ěý
Protein sequence from genomic DNA.
Protein 3D structure from sequence.
Protein function from structure.
Protein function from sequence.
Sequence alignment



Sequence alignmentDr. Harisingh Gour Vishwavidyalaya (A Central Universuty), Sagar, MP
Ěý
This document discusses sequence alignment, which involves arranging biological sequences like DNA, RNA, or proteins to identify regions of similarity. It covers the basic concepts of sequence alignment including global versus local alignment and different methods like dot matrix, dynamic programming, and word-based approaches. Dynamic programming is highlighted as the most common algorithm that uses a scoring system to find the optimal alignment between two sequences.2015 bioinformatics alignments_wim_vancriekinge



2015 bioinformatics alignments_wim_vancriekingeProf. Wim Van Criekinge
Ěý
The document describes an algorithm for pairwise sequence alignment using dynamic programming. It provides an example of applying the algorithm to find the optimal alignment between a zinc-finger protein sequence and a viral protein sequence fragment. The algorithm works by building up the optimal alignment score matrix from left to right and top to bottom, tracking the maximum score at each point to recursively build up to the final alignment.Alignments



AlignmentsJames McInerney
Ěý
The document discusses multiple sequence alignment methods. It describes ClustalW, a commonly used progressive alignment method that first performs pairwise alignments of sequences and constructs a guide tree before progressively aligning sequences based on the tree. ClustalW is fast but has limitations as it is a heuristic that may not find the optimal alignment and provides no way to quantify alignment accuracy.2016 bioinformatics i_alignments_wim_vancriekinge



2016 bioinformatics i_alignments_wim_vancriekingeProf. Wim Van Criekinge
Ěý
- Dynamic programming is used to find the optimal alignment between two protein sequences by recursively computing sub-alignments and storing them in a lookup table.
- The example shows calculating the alignment score between a zinc-finger core sequence and a viral sequence fragment by filling a table and tracking the cumulative scores.
- Filling the table from left to right and top to bottom allows reconstructing the highest scoring alignment between the two sequences.Bioinformatica t3-scoring matrices



Bioinformatica t3-scoring matricesProf. Wim Van Criekinge
Ěý
The document discusses:
1) An overview of bioinformatics lessons including introductions to databases, scoring matrices, and pairwise sequence alignment.
2) Descriptions of major bioinformatics databases and resources including NCBI, ExPASy, and EBI.
3) The importance of scoring matrices in sequence analysis and how the choice of matrix can influence outcomes. Matrices are discussed for nucleotides and proteins.Bioinformatica 20-10-2011-t3-scoring matrices



Bioinformatica 20-10-2011-t3-scoring matricesProf. Wim Van Criekinge
Ěý
This document provides an overview of topics to be covered in a bioinformatics course, including biological databases, sequence similarity scoring matrices, sequence alignments, database searching, phylogenetics, protein structure, gene prediction, and other topics. A schedule is given listing the topics and dates. Background information is also provided on definitions, major bioinformatics databases, scoring matrices, and sequence alignments.Laboratory 1 sequence_alignments



Laboratory 1 sequence_alignmentsseham15
Ěý
This document discusses sequence alignment, which is important for predicting function, database searching, gene finding, and studying sequence divergence. It describes global and local alignment, and algorithms like Needleman-Wunsch, Smith-Waterman, and BLAST that are used for sequence alignment. Sequence alignment finds the best match between sequences and can provide information about molecular evolution by identifying mutations, insertions, and deletions.Sequence alig Sequence Alignment Pairwise alignment:-



Sequence alig Sequence Alignment Pairwise alignment:-naveed ul mushtaq
Ěý
Sequence Alignment Pairwise alignment:-
Global Alignment and Local AlignmentTwo types of alignment Progressive Programs for multiple sequence alignment BLOSUM Point accepted mutation (PAM)PAM VS BLOSUMPart 4 of RNA-seq for DE analysis: Extracting count table and QC



Part 4 of RNA-seq for DE analysis: Extracting count table and QCJoachim Jacob
Ěý
Fourth part of the training session 'RNA-seq for Differential expression analysis'. We explain how we get a count table from a mapping result. We show how to do quality control on the count table. Interested in following this session? Please contact http://www.jakonix.be/contact.htmlIncreasingly Accurate Representation of Biochemistry (v2)



Increasingly Accurate Representation of Biochemistry (v2)Michel Dumontier
Ěý
Biochemical ontologies aim to capture and represent biochemical entities and the relations that exist between them in an accurate manner. A fundamental starting point is biochemical identity, but our current approach for generating identifiers is haphazard and consequently integrating data is error-prone. I will discuss plausible structure-based strategies for biochemical identity whether it be at molecular level or some part thereof (e.g. residues, collection of residues, atoms, collection of atoms, functional groups) such that identifiers may be generated in an automatic and curator/database independent manner. With structure-based identifiers in hand, we will be in a position to more accurately capture context-specific biochemical knowledge, such as how a set of residues in a binding site are involved in a chemical reaction including the fact that a key nitrogen atom must first be de-protonated. Thus, our current representation of biochemical knowledge may improve such that manual and automatic methods of bio-curation are substantially more accurate.Protein Threading



Protein ThreadingSANJANA PANDEY
Ěý
Protein threading is a protein structure prediction method that involves "threading" or placing an amino acid sequence into known protein structure templates to find the best matching fold. The key steps are:
1) A query sequence is threaded into structural positions of templates from a structure library to find sequence-structure alignments
2) Alignments are scored and optimized using an objective function accounting for residue interactions and preferences
3) The highest scoring template is selected as the predicted structure, though loop regions are often not accurately predictedNeedlemanWunsch.pdf



NeedlemanWunsch.pdfYogeshwari54
Ěý
The document describes the Needleman-Wunsch algorithm for global sequence alignment. It discusses sequence comparison and alignment, the scoring scheme including substitution matrices and gap penalties, and provides an overview of how the Needleman-Wunsch algorithm works using dynamic programming to find the optimal global alignment between two sequences.Randomizing genome-scale metabolic networks



Randomizing genome-scale metabolic networksAreejit Samal
Ěý
The document proposes a new Markov Chain Monte Carlo (MCMC) based method to generate randomized metabolic networks that impose biochemical and functional constraints. The method successively constrains the networks by (1) fixing the number of reactions, (2) fixing the number of metabolites, (3) excluding blocked reactions, and (4) requiring growth on a specified environment. Imposing these constraints causes the randomized networks to more closely match properties of real metabolic networks like E. coli. The approach generates an ensemble of diverse yet meaningful randomized networks to help identify design principles in metabolic networks.Msa & rooted/unrooted tree



Msa & rooted/unrooted treeSamiul Ehsan
Ěý
Phylogenetics is the study of evolutionary history and relationships between taxa. Phylogenetic trees present relationships as a collection of nodes and branches, with closely related taxa appearing near each other. Multiple sequence alignment (MSA) is used to reveal biological facts about sequences and to construct phylogenetic trees. However, MSA is computationally complex due to the exponential growth in possible alignments as more sequences are added.Sequence Alignment



Sequence AlignmentRavi Gandham
Ěý
Sequence alignment involves arranging DNA, RNA, or protein sequences to identify regions of similarity. It is used to determine if sequences are evolutionarily related, observe patterns of conservation, and find similar regions within proteins. The key steps are representation of sequences in a matrix, insertion of gaps, and use of scoring schemes like PAM and BLOSUM matrices to identify the best alignment. Global alignment forces alignment over full sequence lengths while local alignment identifies short, well-matching segments. Algorithms like Needleman-Wunsch and Smith-Waterman use dynamic programming to calculate optimal pairwise sequence alignments.Yale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...



Yale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...Yale School of Public Health - The Virtual Medical Operations Center (VMOC)
Ěý
Measles Outbreak—Southwestern US— This briefing reviews the current situation surrounding the measles outbreaks in Texas, New Mexico, Oklahoma, and Kansas. Role of Teacher in the era of Generative AI



Role of Teacher in the era of Generative AIProf. Neeta Awasthy
Ěý
We need to layer the technology onto existing workflows
Follow the teachers who inspire you because that instills passion Curiosity & Lifelong Learning.
You can benefit from generative AI even when its intelligence is worse-because of the potential for cost and time savings in low-cost-of-error environments.
Bot tutors are already yielding effective results on learning and mastery.
GenAI may increase the digital divide- its gains may accrue disproportionately to those who already have domain expertise.
GenAI can be used for Coding
Complex structures
Make the content
Manage the content
Solutions to complex numerical problems
Lesson plan
Assignment
Quiz
Question bank
Report & summary of content
Creating videos
Title of abstract & summaries and much more like...
Improving Grant Writing
Learning by Teaching Chatbots
GenAI as peer Learner
Data Analysis for Non-Coders
Student Course Preparation
To reduce Plagiarism
Legal Problems for classes
Understanding Student Learning in Real Time
Simulate a poor
Faculty co-pilot chatbot
Generate fresh Assessments
Data Analysis Partner
Summarize student questions in real-time
Assess depth of students' understanding
The skills to foster are Listening
Communicating
Approaching the problem & solving
Making Real Time Decisions
Logic
Refining Memories
Learning Cultures & Syntax (Foreign Language)
Chatbots & Agentic AI can never so what a professor can do.
The need of the hour is to teach Creativity
Emotions
Judgement
Psychology
Communication
Human Emotions
…………Through various content!
More Related Content
Similar to Sequence Alignment - Data Bioinformatics Introduction (20)
An Efficient Biological Sequence Compression Technique Using LUT and Repeat ...



An Efficient Biological Sequence Compression Technique Using LUT and Repeat ...IOSR Journals
Ěý
This document presents two improved biological sequence compression algorithms that utilize a lookup table (LUT) and identification of tandem repeats in sequences. The first algorithm maps all possible 3-character combinations to ASCII characters using a 125-entry LUT. The second maps all possible 4-character combinations to ASCII characters using a 256-entry LUT. These algorithms aim to achieve high compression factors, saving percentages, and faster compression/decompression times compared to previous biological sequence compression methods.DNA Compression (Encoded using Huffman Encoding Method)



DNA Compression (Encoded using Huffman Encoding Method)Marwa Al-Rikaby
Ěý
The document discusses DNA compression algorithms. It describes the common components of most DNA compression algorithms, which include finding repeat segments in a DNA sequence, considering approximate repeats allowing for operations like substitutions, and selecting the best set of compatible repeats. It then provides an example demonstrating how these steps may be applied to a sample DNA sequence to identify repeat segments and encode them for compression.lecture4.ppt Sequence Alignmentaldf sdfsadf



lecture4.ppt Sequence Alignmentaldf sdfsadfalizain9604
Ěý
This document summarizes key concepts in sequence alignment including:
1) Sequence alignment involves finding the linear correspondence between symbols in one sequence to another that maximizes similarity. Dynamic programming is commonly used to compute optimal alignments.
2) BLAST is an extremely fast database search tool that uses heuristics like word matching to find local alignments and statistical analysis to assess significance.
3) Multiple sequence alignments make conserved features more apparent but are more difficult to compute than pairwise alignments. Progressive alignment gradually merges pairwise alignments based on a phylogenetic tree.Basics of bioinformatics



Basics of bioinformaticsAbhishek Vatsa
Ěý
Bioinformatics emerged from the marriage of computer science and molecular biology to analyze massive amounts of biological data, like that produced by the Human Genome Project. It uses algorithms and techniques from computer science to solve problems in molecular biology, like comparing genomic sequences to understand evolution. As genomic data exploded publicly, bioinformatics was needed to efficiently store, analyze, and make sense of this information, which has applications in molecular medicine, drug development, agriculture, and more.protein structure prediction in bioinformatics.ppt



protein structure prediction in bioinformatics.pptDrSudha2
Ěý
Protein sequence from genomic DNA.
Protein 3D structure from sequence.
Protein function from structure.
Protein function from sequence.
Sequence alignment



Sequence alignmentDr. Harisingh Gour Vishwavidyalaya (A Central Universuty), Sagar, MP
Ěý
This document discusses sequence alignment, which involves arranging biological sequences like DNA, RNA, or proteins to identify regions of similarity. It covers the basic concepts of sequence alignment including global versus local alignment and different methods like dot matrix, dynamic programming, and word-based approaches. Dynamic programming is highlighted as the most common algorithm that uses a scoring system to find the optimal alignment between two sequences.2015 bioinformatics alignments_wim_vancriekinge



2015 bioinformatics alignments_wim_vancriekingeProf. Wim Van Criekinge
Ěý
The document describes an algorithm for pairwise sequence alignment using dynamic programming. It provides an example of applying the algorithm to find the optimal alignment between a zinc-finger protein sequence and a viral protein sequence fragment. The algorithm works by building up the optimal alignment score matrix from left to right and top to bottom, tracking the maximum score at each point to recursively build up to the final alignment.Alignments



AlignmentsJames McInerney
Ěý
The document discusses multiple sequence alignment methods. It describes ClustalW, a commonly used progressive alignment method that first performs pairwise alignments of sequences and constructs a guide tree before progressively aligning sequences based on the tree. ClustalW is fast but has limitations as it is a heuristic that may not find the optimal alignment and provides no way to quantify alignment accuracy.2016 bioinformatics i_alignments_wim_vancriekinge



2016 bioinformatics i_alignments_wim_vancriekingeProf. Wim Van Criekinge
Ěý
- Dynamic programming is used to find the optimal alignment between two protein sequences by recursively computing sub-alignments and storing them in a lookup table.
- The example shows calculating the alignment score between a zinc-finger core sequence and a viral sequence fragment by filling a table and tracking the cumulative scores.
- Filling the table from left to right and top to bottom allows reconstructing the highest scoring alignment between the two sequences.Bioinformatica t3-scoring matrices



Bioinformatica t3-scoring matricesProf. Wim Van Criekinge
Ěý
The document discusses:
1) An overview of bioinformatics lessons including introductions to databases, scoring matrices, and pairwise sequence alignment.
2) Descriptions of major bioinformatics databases and resources including NCBI, ExPASy, and EBI.
3) The importance of scoring matrices in sequence analysis and how the choice of matrix can influence outcomes. Matrices are discussed for nucleotides and proteins.Bioinformatica 20-10-2011-t3-scoring matrices



Bioinformatica 20-10-2011-t3-scoring matricesProf. Wim Van Criekinge
Ěý
This document provides an overview of topics to be covered in a bioinformatics course, including biological databases, sequence similarity scoring matrices, sequence alignments, database searching, phylogenetics, protein structure, gene prediction, and other topics. A schedule is given listing the topics and dates. Background information is also provided on definitions, major bioinformatics databases, scoring matrices, and sequence alignments.Laboratory 1 sequence_alignments



Laboratory 1 sequence_alignmentsseham15
Ěý
This document discusses sequence alignment, which is important for predicting function, database searching, gene finding, and studying sequence divergence. It describes global and local alignment, and algorithms like Needleman-Wunsch, Smith-Waterman, and BLAST that are used for sequence alignment. Sequence alignment finds the best match between sequences and can provide information about molecular evolution by identifying mutations, insertions, and deletions.Sequence alig Sequence Alignment Pairwise alignment:-



Sequence alig Sequence Alignment Pairwise alignment:-naveed ul mushtaq
Ěý
Sequence Alignment Pairwise alignment:-
Global Alignment and Local AlignmentTwo types of alignment Progressive Programs for multiple sequence alignment BLOSUM Point accepted mutation (PAM)PAM VS BLOSUMPart 4 of RNA-seq for DE analysis: Extracting count table and QC



Part 4 of RNA-seq for DE analysis: Extracting count table and QCJoachim Jacob
Ěý
Fourth part of the training session 'RNA-seq for Differential expression analysis'. We explain how we get a count table from a mapping result. We show how to do quality control on the count table. Interested in following this session? Please contact http://www.jakonix.be/contact.htmlIncreasingly Accurate Representation of Biochemistry (v2)



Increasingly Accurate Representation of Biochemistry (v2)Michel Dumontier
Ěý
Biochemical ontologies aim to capture and represent biochemical entities and the relations that exist between them in an accurate manner. A fundamental starting point is biochemical identity, but our current approach for generating identifiers is haphazard and consequently integrating data is error-prone. I will discuss plausible structure-based strategies for biochemical identity whether it be at molecular level or some part thereof (e.g. residues, collection of residues, atoms, collection of atoms, functional groups) such that identifiers may be generated in an automatic and curator/database independent manner. With structure-based identifiers in hand, we will be in a position to more accurately capture context-specific biochemical knowledge, such as how a set of residues in a binding site are involved in a chemical reaction including the fact that a key nitrogen atom must first be de-protonated. Thus, our current representation of biochemical knowledge may improve such that manual and automatic methods of bio-curation are substantially more accurate.Protein Threading



Protein ThreadingSANJANA PANDEY
Ěý
Protein threading is a protein structure prediction method that involves "threading" or placing an amino acid sequence into known protein structure templates to find the best matching fold. The key steps are:
1) A query sequence is threaded into structural positions of templates from a structure library to find sequence-structure alignments
2) Alignments are scored and optimized using an objective function accounting for residue interactions and preferences
3) The highest scoring template is selected as the predicted structure, though loop regions are often not accurately predictedNeedlemanWunsch.pdf



NeedlemanWunsch.pdfYogeshwari54
Ěý
The document describes the Needleman-Wunsch algorithm for global sequence alignment. It discusses sequence comparison and alignment, the scoring scheme including substitution matrices and gap penalties, and provides an overview of how the Needleman-Wunsch algorithm works using dynamic programming to find the optimal global alignment between two sequences.Randomizing genome-scale metabolic networks



Randomizing genome-scale metabolic networksAreejit Samal
Ěý
The document proposes a new Markov Chain Monte Carlo (MCMC) based method to generate randomized metabolic networks that impose biochemical and functional constraints. The method successively constrains the networks by (1) fixing the number of reactions, (2) fixing the number of metabolites, (3) excluding blocked reactions, and (4) requiring growth on a specified environment. Imposing these constraints causes the randomized networks to more closely match properties of real metabolic networks like E. coli. The approach generates an ensemble of diverse yet meaningful randomized networks to help identify design principles in metabolic networks.Msa & rooted/unrooted tree



Msa & rooted/unrooted treeSamiul Ehsan
Ěý
Phylogenetics is the study of evolutionary history and relationships between taxa. Phylogenetic trees present relationships as a collection of nodes and branches, with closely related taxa appearing near each other. Multiple sequence alignment (MSA) is used to reveal biological facts about sequences and to construct phylogenetic trees. However, MSA is computationally complex due to the exponential growth in possible alignments as more sequences are added.Sequence Alignment



Sequence AlignmentRavi Gandham
Ěý
Sequence alignment involves arranging DNA, RNA, or protein sequences to identify regions of similarity. It is used to determine if sequences are evolutionarily related, observe patterns of conservation, and find similar regions within proteins. The key steps are representation of sequences in a matrix, insertion of gaps, and use of scoring schemes like PAM and BLOSUM matrices to identify the best alignment. Global alignment forces alignment over full sequence lengths while local alignment identifies short, well-matching segments. Algorithms like Needleman-Wunsch and Smith-Waterman use dynamic programming to calculate optimal pairwise sequence alignments.Recently uploaded (20)
Yale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...



Yale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...Yale School of Public Health - The Virtual Medical Operations Center (VMOC)
Ěý
Measles Outbreak—Southwestern US— This briefing reviews the current situation surrounding the measles outbreaks in Texas, New Mexico, Oklahoma, and Kansas. Role of Teacher in the era of Generative AI



Role of Teacher in the era of Generative AIProf. Neeta Awasthy
Ěý
We need to layer the technology onto existing workflows
Follow the teachers who inspire you because that instills passion Curiosity & Lifelong Learning.
You can benefit from generative AI even when its intelligence is worse-because of the potential for cost and time savings in low-cost-of-error environments.
Bot tutors are already yielding effective results on learning and mastery.
GenAI may increase the digital divide- its gains may accrue disproportionately to those who already have domain expertise.
GenAI can be used for Coding
Complex structures
Make the content
Manage the content
Solutions to complex numerical problems
Lesson plan
Assignment
Quiz
Question bank
Report & summary of content
Creating videos
Title of abstract & summaries and much more like...
Improving Grant Writing
Learning by Teaching Chatbots
GenAI as peer Learner
Data Analysis for Non-Coders
Student Course Preparation
To reduce Plagiarism
Legal Problems for classes
Understanding Student Learning in Real Time
Simulate a poor
Faculty co-pilot chatbot
Generate fresh Assessments
Data Analysis Partner
Summarize student questions in real-time
Assess depth of students' understanding
The skills to foster are Listening
Communicating
Approaching the problem & solving
Making Real Time Decisions
Logic
Refining Memories
Learning Cultures & Syntax (Foreign Language)
Chatbots & Agentic AI can never so what a professor can do.
The need of the hour is to teach Creativity
Emotions
Judgement
Psychology
Communication
Human Emotions
…………Through various content!
Viceroys of India & Their Tenure – Key Events During British Rule



Viceroys of India & Their Tenure – Key Events During British RuleDeeptiKumari61
Ěý
The British Raj in India (1857-1947) saw significant events under various Viceroys, shaping the political, economic, and social landscape.
**Early Period (1856-1888):**
Lord Canning (1856-1862) handled the Revolt of 1857, leading to the British Crown taking direct control. Universities were established, and the Indian Councils Act (1861) was passed. Lord Lawrence (1864-1869) led the Bhutan War and established High Courts. Lord Lytton (1876-1880) enforced repressive laws like the Vernacular Press Act (1878) and Arms Act (1878) while waging the Second Afghan War.
**Reforms & Political Awakening (1880-1905):**
Lord Ripon (1880-1884) introduced the Factory Act (1881), Local Self-Government Resolution (1882), and repealed the Vernacular Press Act. Lord Dufferin (1884-1888) oversaw the formation of the Indian National Congress (1885). Lord Lansdowne (1888-1894) passed the Factory Act (1891) and Indian Councils Act (1892). Lord Curzon (1899-1905) introduced educational reforms but faced backlash for the Partition of Bengal (1905).
**Rise of Nationalism (1905-1931):**
Lord Minto II (1905-1910) saw the rise of the Swadeshi Movement and the Muslim League's formation (1906). Lord Hardinge II (1910-1916) annulled Bengal’s Partition (1911) and shifted India’s capital to Delhi. Lord Chelmsford (1916-1921) faced the Lucknow Pact (1916), Jallianwala Bagh Massacre (1919), and Non-Cooperation Movement. Lord Reading (1921-1926) dealt with the Chauri Chaura Incident (1922) and the formation of the Swaraj Party. Lord Irwin (1926-1931) saw the Simon Commission protests, the Dandi March, and the Gandhi-Irwin Pact (1931).
**Towards Independence (1931-1947):**
Lord Willingdon (1931-1936) introduced the Government of India Act (1935), laying India's federal framework. Lord Linlithgow (1936-1944) faced WWII-related crises, including the Quit India Movement (1942). Lord Wavell (1944-1947) proposed the Cabinet Mission Plan (1946) and negotiated British withdrawal. Lord Mountbatten (1947-1948) oversaw India's Partition and Independence on August 15, 1947.
**Final Transition:**
C. Rajagopalachari (1948-1950), India’s last Governor-General, facilitated India’s transition into a republic before the position was abolished in 1950.
The British Viceroys played a crucial role in India’s colonial history, introducing both repressive and progressive policies that fueled nationalist movements, ultimately leading to independence.https://www.youtube.com/@DKDEducationHow to Configure Outgoing and Incoming mail servers in Odoo 18



How to Configure Outgoing and Incoming mail servers in Odoo 18Celine George
Ěý
Odoo 18 features a powerful email management system designed to streamline business communications directly within the platform. By setting up Outgoing Mail Servers, users can effortlessly send emails. Similarly, configuring Incoming Mail Servers enables Odoo to process incoming emails and generate records such as leads or helpdesk tickets. Key Frameworks in Systematic Reviews - Dr Reginald Quansah



Key Frameworks in Systematic Reviews - Dr Reginald QuansahSystematic Reviews Network (SRN)
Ěý
Recognize features of systematic reviews and meta-analyses as a research design
Identify the elements of a well-defined review question
Understand and develop search strategies and able to turn research questions into search strategy
Perform a comprehensive search for relevant studies
Manage the results of systematic searches
Extract data and assess risk of bias of included studies
Understand and carry out quantitative analysis of extracted data
Apply the methodology and conduct reviews independentlyAnti-Viral Agents.pptx Medicinal Chemistry III, B Pharm SEM VI



Anti-Viral Agents.pptx Medicinal Chemistry III, B Pharm SEM VISamruddhi Khonde
Ěý
Antiviral agents are crucial in combating viral infections, causing a variety of diseases from mild to life-threatening. Developed through medicinal chemistry, these drugs target viral structures and processes while minimizing harm to host cells. Viruses are classified into DNA and RNA viruses, with each replicating through distinct mechanisms. Treatments for herpesviruses involve nucleoside analogs like acyclovir and valacyclovir, which inhibit the viral DNA polymerase. Influenza is managed with neuraminidase inhibitors like oseltamivir and zanamivir, which prevent the release of new viral particles. HIV is treated with a combination of antiretroviral drugs targeting various stages of the viral life cycle. Hepatitis B and C are treated with different strategies, with nucleoside analogs like lamivudine inhibiting viral replication and direct-acting antivirals targeting the viral RNA polymerase and other key proteins.
Antiviral agents are designed based on their mechanisms of action, with several categories including nucleoside and nucleotide analogs, protease inhibitors, neuraminidase inhibitors, reverse transcriptase inhibitors, and integrase inhibitors. The design of these agents often relies on understanding the structure-activity relationship (SAR), which involves modifying the chemical structure of compounds to enhance efficacy, selectivity, and bioavailability while reducing side effects. Despite their success, challenges such as drug resistance, viral mutation, and the need for long-term therapy remain.How to Manage Purchase Order Approval in Odoo 18



How to Manage Purchase Order Approval in Odoo 18Celine George
Ěý
In Odoo 18, you can set a minimum amount as a limit, and whenever an order comes above the limit, it requires the approval of the manager. In this slide, we are diving into the crucial aspect of procurement, which is managing purchase order approval. MICROECONOMICS: RENT AND THEORIES OF RENT



MICROECONOMICS: RENT AND THEORIES OF RENTDrSundariD
Ěý
CONCEPTS COVERED: RENT, THEORIES OF RENT, QUASI RENTHow to Setup Company Data in Odoo 17 Accounting App



How to Setup Company Data in Odoo 17 Accounting AppCeline George
Ěý
The Accounting module in Odoo 17 is a comprehensive tool designed to manage all financial aspects of a business. It provides a range of features that help with everything from day-to-day bookkeeping to advanced financial analysis. Knownsense 2025 prelims- U-25 General Quiz.pdf



Knownsense 2025 prelims- U-25 General Quiz.pdfPragya - UEM Kolkata Quiz Club
Ěý
General College Quiz conducted by Pragya the Official Quiz Club of the University of Engineering and Management Kolkata in collaboration with Ecstasia the official cultural fest of the University of Engineering and Management Kolkata.Anti-Fungal Agents.pptx Medicinal Chemistry III B. Pharm Sem VI



Anti-Fungal Agents.pptx Medicinal Chemistry III B. Pharm Sem VISamruddhi Khonde
Ěý
UNIT – IV
Antifungal agents:
Antifungal antibiotics: Amphotericin-B, Nystatin, Natamycin, Griseofulvin.
Synthetic Antifungal agents: Clotrimazole, Econazole, Butoconazole,
Oxiconazole Tioconozole, Miconazole*, Ketoconazole, Terconazole,
Itraconazole, Fluconazole, Naftifine hydrochloride, Tolnaftate*Antifungal agents by Mrs. Manjushri Dabhade



Antifungal agents by Mrs. Manjushri DabhadeDabhade madam Dabhade
Ěý
Information about chemical structure, MOA, ClassificationUTI Quinolones by Mrs. Manjushri Dabhade



UTI Quinolones by Mrs. Manjushri DabhadeDabhade madam Dabhade
Ěý
UTI quinolones by Mrs. Manjushri Dabhade3. AI Trust Layer, Governance – Explainability, Security & Compliance.pdf



3. AI Trust Layer, Governance – Explainability, Security & Compliance.pdfMukesh Kala
Ěý
AI Trust Layer, Governance – Explainability, Security & ComplianceUnit1 Inroduction to Internal Combustion Engines



Unit1 Inroduction to Internal Combustion EnginesNileshKumbhar21
Ěý
Introduction of I. C. Engines, Types of engine, working of engine, Nomenclature of engine, Otto cycle, Diesel cycle Fuel air cycles Characteristics of fuel - air mixtures Actual cycles, Valve timing diagram for high and low speed engine, Port timing diagramYale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...



Yale VMOC Special Report - Measles Outbreak Southwest US 3-30-2025 FINAL v2...Yale School of Public Health - The Virtual Medical Operations Center (VMOC)
Ěý
Sequence Alignment - Data Bioinformatics Introduction
- 1. Learning from primary structure Sequence alignment
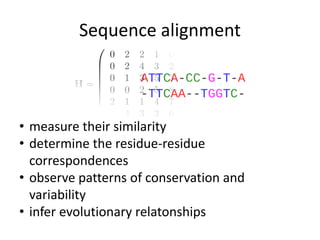
- 2. Sequence alignment • measure their similarity • determine the residue-residue correspondences • observe patterns of conservation and variability • infer evolutionary relatonships
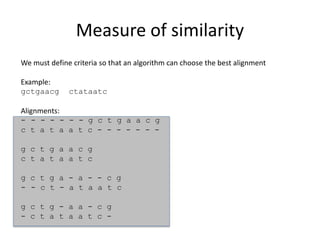
- 3. Measure of similarity alignment: identification of residue-residue correspondences Correspondences must preserve the order of residues Gaps may be introduced Example: First string= a b c d e second string= a c d e f A reasonable alignment: a b c d e – a – c d e f
- 4. Measure of similarity We must define criteria so that an algorithm can choose the best alignment Example: gctgaacg ctataatc Alignments: - - - - - - - g c t g a a c g c t a t a a t c - - - - - - - g c t g a a c g c t a t a a t c g c t g a - a - - c g - - c t - a t a a t c g c t g - a a - c g - c t a t a a t c -
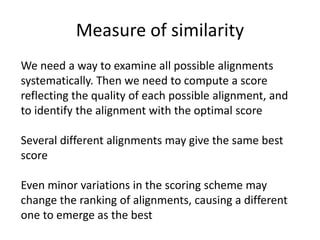
- 5. Measure of similarity We need a way to examine all possible alignments systematically. Then we need to compute a score reflecting the quality of each possible alignment, and to identify the alignment with the optimal score Several different alignments may give the same best score Even minor variations in the scoring scheme may change the ranking of alignments, causing a different one to emerge as the best
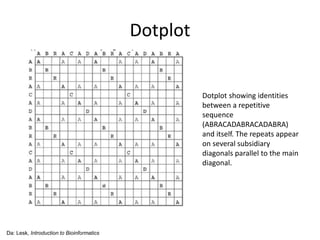
- 6. Dotplot • give an overview of the similarities between two sequences • have a close relationship with the alignment between two sequences Da: Lesk, Introduction to Bioinformatics Dotplot showing identities between short name (DOROTHYHODGKIN) and full name (DOROTHYCROWFOOTH ODGKIN)
- 7. Dotplot Da: Lesk, Introduction to Bioinformatics Dotplot showing identities between a repetitive sequence (ABRACADABRACADABRA) and itself. The repeats appear on several subsidiary diagonals parallel to the main diagonal.
- 8. Dotplot Da: Lesk, Introduction to Bioinformatics Dotplot showing identities between the palindromic sequence MAX I STAY AWAY AT SIX AM and itself. The palindrome reveals itself as a stretch of matches perpendicular to the main diagonal Remember that: Restriction enzymes and transcriptional regulatory factors may recognize palindrome sequences EcoRI: GAATTC CTTAAG
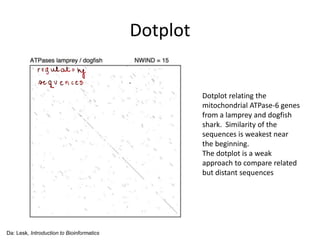
- 9. Dotplot Da: Lesk, Introduction to Bioinformatics Dotplot relating the mitochondrial ATPase-6 genes from a lamprey and dogfish shark. Similarity of the sequences is weakest near the beginning. The dotplot is a weak approach to compare related but distant sequences
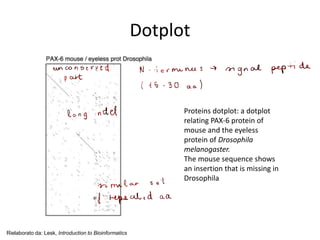
- 10. Dotplot Proteins dotplot: a dotplot relating PAX-6 protein of mouse and the eyeless protein of Drosophila melanogaster. The mouse sequence shows an insertion that is missing in Drosophila Rielaborato da: Lesk, Introduction to Bioinformatics
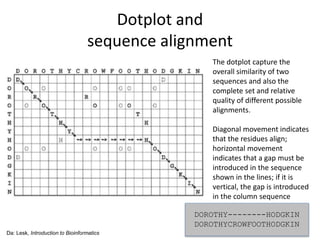
- 11. Dotplot and sequence alignment The dotplot capture the overall similarity of two sequences and also the complete set and relative quality of different possible alignments. Diagonal movement indicates that the residues align; horizontal movement indicates that a gap must be introduced in the sequence shown in the lines; if it is vertical, the gap is introduced in the column sequence Da: Lesk, Introduction to Bioinformatics DOROTHY--------HODGKIN DOROTHYCROWFOOTHODGKIN
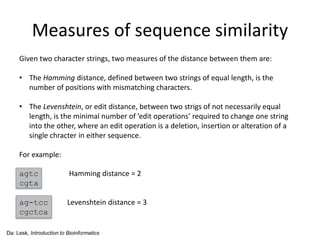
- 12. Measures of sequence similarity Given two character strings, two measures of the distance between them are: • The Hamming distance, defined between two strings of equal length, is the number of positions with mismatching characters. • The Levenshtein, or edit distance, between two strigs of not necessarily equal length, is the minimal number of ’edit operations’ required to change one string into the other, where an edit operation is a deletion, insertion or alteration of a single chracter in either sequence. For example: agtc Hamming distance = 2 cgta ag-tcc Levenshtein distance = 3 cgctca Da: Lesk, Introduction to Bioinformatics




