20141105 古代ゲノムジャーナルクラブ
- 1. SI 20 “Expression pattern enrichment among genes with protein coding changes in humans” 2014/11/05 古代ゲノムJC in 三島 統計数理研究所 松前ひろみ 14年11月5日水曜日
- 2. このセクションが試みていること ? Homo sapiensとは何か=現代人の知能を、遺伝子レベルで意味づけをしたい ? ヒトおよび霊長類の脳の時空間特異的遺伝子発現データに、今回見つかって きた現代人特異的な遺伝子群(変異)をマッピング ? 脳の中で、ヒトらしさを生んでいる遺伝子群を同定したい 14年11月5日水曜日
- 3. 14年11月5日水曜日
- 4. Allen Institute for Brain Science ? マイクロソフトの共同創業者のPaul Allenが2003に1億ドルを出資して設立した脳科学の研究所 ? Research Science ? Structured Science ? Allen Brain Atlas (Hawrylycz et al. 2012, Nature)の公開で有名 ? http://www.brain-map.org/ Paul Allen ? ヒト、マウス、霊長類 ? 脳2D/3Dマップ ? 解剖学的部位 ? 発生過程 ? イメージング ? 遺伝子発現 14年11月5日水曜日
- 5. ? 現代人のみで ? コーディング領域に置換が起きている 87の遺伝子 ? 脳のヒト特異的な領域や、発生過程 の発現に関わる遺伝子 ? 同義置換が起きている108の遺伝子 14年11月5日水曜日
- 6. 脳に関わる遺伝子の収集”enrichment” ? 脳の遺伝子発現解析は難しいことが知られている ? 死後の組織しか採取できない ? RNAが分解しやすい ? controlが難しい ? 脳の時空間データベース ? Human Brain Atlas (adult) (Hawrylycz et al. 2012, Nature) ? BrainSpan Atlas of the Developing Human Brain (Kang et al. 2011, Nature) ? http://www.brainspan.org/ ? 皮質の形成に重要な時期(15-20週)のデータも含む ? マイクロアレイの遺伝子発現データ ? NIH Blueprint Non-Human Primate Atlas (Rhesus macaque) ? http://www.blueprintnhpatlas.org/macrodissection/index ? adult macaque monkey microarray atlas ? http://www.blueprintnhpatlas.org/nhp/download.html 14年11月5日水曜日
- 7. NIH Blueprint Non-Human Primate Atlas (Rhesus macaque) 14年11月5日水曜日
- 8. 時空間的発現パターンを抽出する ? “遺伝子群の機能単位(パスウェイ)”で発現パターンが似ているはず? ? 発現しているからと言って機能している訳ではないが、同じ機能に関わっている遺伝子群が共発現してい ることに意味があるのではないか? ? 共発現遺伝子ネットワークの抽出, Pathway Enrichment Analysis ? Weighted Gene Co-expression Network Analysis (WGCNA) (Langfelder et al. 2008, BMC Bioinformatics; R package) ? 現代人特異的に見つかってきた、非同義置換?同義置換の2つの遺伝子セットの間に、脳の中で部位?時間特 異的な共発現ネットワークが見つかるか? 14年11月5日水曜日
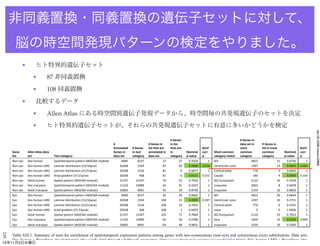
- 9. 248 # Genes # Genes in # # Genes in in list data set in # Genes in Annotated # Genes list that are that are Bonf most list in most Bonf Gene Allen Atlas data Genes in in test annotated in in Nominal corr Most common common common Nominal corr list set Test Category data set category data set category p-value p category match category category p-value p Non-syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 37 37 0.3329 1 M1 4655 21 0.4756 1 Non-syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 81 20 0.0006 0.016 Ventricular zone 1497 14 0.0023 0.064 Non-syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 81 9 0.5877 1 Cortical plate 776 3 0.6042 1 Non-syn Dev human LMD Areal gradient (15-21pcw) 20268 468 81 6 0.0111 0.311 Caudal 180 4 0.0059 0.165 Non-syn Adult human Spatial pattern (WGCNA module) 21337 15347 76 52 0.7926 1 M2 (turquoise) 5110 16 0.7630 1 Non-syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 44 41 0.4207 1 turquoise 2062 8 0.5878 1 Non-syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 43 34 0.8740 1 turquoise 3154 10 0.8603 1 Syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 42 39 0.9665 1 M1 4655 20 0.8644 1 Syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 108 21 0.0095 0.267 Ventricular zone 1497 10 0.2751 1 Syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 108 14 0.3501 1 Cortical plate 776 6 0.2331 1 Syn Dev human LMD Areal gradient (15-21pcw) 20268 468 108 1 0.9203 1 none 0 0 1.0000 1 Syn Adult human Spatial pattern (WGCNA module) 21337 15347 103 71 0.7864 1 M2 (turquoise) 5110 22 0.7653 1 Syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 69 58 0.9788 1 blue 1609 16 0.0354 0.990 Syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 64 48 0.9825 1 turquoise 3154 19 0.5393 1 Table S20.1. Summary of tests for enrichment of spatiotemporal expression patterns among genes with non-synonymous (non-syn) and synonymous (syn) substitutions. Data sets: Dev human = BrainSpan developmental atlas (early fetal through adulthood) microarray (http://www.brainspan.org/rnaseq/search/index.html); Dev human LMD = BrainSpan atlas doi:10.1038/nature 非同義置換?同義置換の遺伝子セットに対して、 脳の時空間発現パターンの検定をやりました。 ? ヒト特異的遺伝子セット ? 87 非同義置換 ? 108 同義置換 ? 比較するデータ ? Allen Atlas にある時空間別遺伝子発現データから、時空間毎の共発現遺伝子のセットを決定 ? ヒト特異的遺伝子セットが、それらの共発現遺伝子セットに有意に多いかどうかを検定 14年11月5日水曜日
- 10. 248 # Genes # Genes in # # Genes in in list data set in # Genes in Annotated # Genes list that are that are Bonf most list in most Bonf Gene Allen Atlas data Genes in in test annotated in in Nominal corr Most common common common Nominal corr list set Test Category data set category data set category p-value p category match category category p-value p Non-syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 37 37 0.3329 1 M1 4655 21 0.4756 1 Non-syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 81 20 0.0006 0.016 Ventricular zone 1497 14 0.0023 0.064 Non-syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 81 9 0.5877 1 Cortical plate 776 3 0.6042 1 Non-syn Dev human LMD Areal gradient (15-21pcw) 20268 468 81 6 0.0111 0.311 Caudal 180 4 0.0059 0.165 Non-syn Adult human Spatial pattern (WGCNA module) 21337 15347 76 52 0.7926 1 M2 (turquoise) 5110 16 0.7630 1 Non-syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 44 41 0.4207 1 turquoise 2062 8 0.5878 1 Non-syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 43 34 0.8740 1 turquoise 3154 10 0.8603 1 Syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 42 39 0.9665 1 M1 4655 20 0.8644 1 Syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 108 21 0.0095 0.267 Ventricular zone 1497 10 0.2751 1 Syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 108 14 0.3501 1 Cortical plate 776 6 0.2331 1 Syn Dev human LMD Areal gradient (15-21pcw) 20268 468 108 1 0.9203 1 none 0 0 1.0000 1 Syn Adult human Spatial pattern (WGCNA module) 21337 15347 103 71 0.7864 1 M2 (turquoise) 5110 22 0.7653 1 Syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 69 58 0.9788 1 blue 1609 16 0.0354 0.990 Syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 64 48 0.9825 1 turquoise 3154 19 0.5393 1 Table S20.1. Summary of tests for enrichment of spatiotemporal expression patterns among genes with non-synonymous (non-syn) and synonymous (syn) substitutions. Data sets: Dev human = BrainSpan developmental atlas (early fetal through adulthood) microarray (http://www.brainspan.org/rnaseq/search/index.html); Dev human LMD = BrainSpan atlas doi:10.1038/nature 非同義置換?同義置換の遺伝子セットに対して、 脳の時空間発現パターンの検定をこれだけやりました。 ? 非同義置換遺伝子群で、Ventricular Zone (15-16週 の大脳皮質の形成期)で発現している遺伝子群に (全遺伝子群と比較して)差があった ? 同義置換遺伝子群では、このカテゴリーには有 意差なし 14年11月5日水曜日
- 11. Fig. S20 大脳皮質形成期の遺伝子発現 random: 新皮質の発生で発現している遺伝子100個を1000回ランダムに選んで平均発現 量を取得 14年11月5日水曜日
- 12. 248 # Genes # Genes in # # Genes in in list data set in # Genes in Annotated # Genes list that are that are Bonf most list in most Bonf Gene Allen Atlas data Genes in in test annotated in in Nominal corr Most common common common Nominal corr list set Test Category data set category data set category p-value p category match category category p-value p Non-syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 37 37 0.3329 1 M1 4655 21 0.4756 1 Non-syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 81 20 0.0006 0.016 Ventricular zone 1497 14 0.0023 0.064 Non-syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 81 9 0.5877 1 Cortical plate 776 3 0.6042 1 Non-syn Dev human LMD Areal gradient (15-21pcw) 20268 468 81 6 0.0111 0.311 Caudal 180 4 0.0059 0.165 Non-syn Adult human Spatial pattern (WGCNA module) 21337 15347 76 52 0.7926 1 M2 (turquoise) 5110 16 0.7630 1 Non-syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 44 41 0.4207 1 turquoise 2062 8 0.5878 1 Non-syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 43 34 0.8740 1 turquoise 3154 10 0.8603 1 Syn Dev human Spatiotemporal pattern (WGCNA module) 8485 8237 42 39 0.9665 1 M1 4655 20 0.8644 1 Syn Dev human LMD Laminar distribution (15/16pcw) 20268 2304 108 21 0.0095 0.267 Ventricular zone 1497 10 0.2751 1 Syn Dev human LMD Laminar distribution (21/21pcw) 20268 2316 108 14 0.3501 1 Cortical plate 776 6 0.2331 1 Syn Dev human LMD Areal gradient (15-21pcw) 20268 468 108 1 0.9203 1 none 0 0 1.0000 1 Syn Adult human Spatial pattern (WGCNA module) 21337 15347 103 71 0.7864 1 M2 (turquoise) 5110 22 0.7653 1 Syn Dev macaque Spatiotemporal pattern (WGCNA module) 11102 10089 69 58 0.9788 1 blue 1609 16 0.0354 0.990 Syn Adult macaque Spatial pattern (WGCNA module) 10665 8991 64 48 0.9825 1 turquoise 3154 19 0.5393 1 Table S20.1. Summary of tests for enrichment of spatiotemporal expression patterns among genes with non-synonymous (non-syn) and synonymous (syn) substitutions. Data sets: Dev human = BrainSpan developmental atlas (early fetal through adulthood) microarray (http://www.brainspan.org/rnaseq/search/index.html); Dev human LMD = BrainSpan atlas doi:10.1038/nature 非同義置換?同義置換の遺伝子セットに対して、 脳の時空間発現パターンの検定をこれだけやりました。 ? 非同義置換:数は少ないが、大脳皮質の発生で発現していて、前頭側頭部?で発現してい る遺伝子群もあった ? SLITRK1(神経突起伸張に関与し、チック症に関連), TKTL1, C21orf62, GLDC, IFI44L, ZNF185 ? (ここにはないけれど)CASC5は大脳皮質形成期に発現して細胞周期に関与し、さ らには小頭症に関与 ? 大脳皮質形成期がヒトらしさを生んでいる? 14年11月5日水曜日