RNAseqによる変動遺伝子抽出の統計: A Review
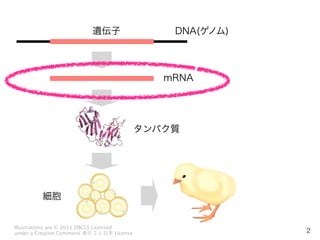
- 2. 2 DNA(ゲノム) mRNA 遺伝子 タンパク質 細胞 Illustrations are ? 2011 DBCLS Licensed under a Creative Commons 表示 2.1 日本 License
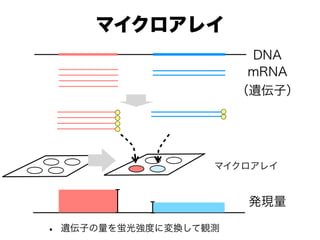
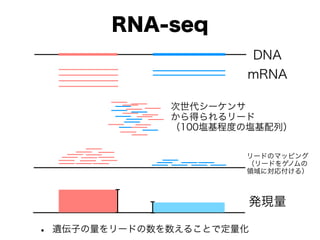
- 3. 大規模発現量観測の小史 各遺伝子由来のタグの計数 ランダムに抽出したmRNA中の,特定の部位の配列を特定し,遺伝子発 現量の推定を行う方法 SAGE [Velculescu et al. 1995], BodyMap [Kawamoto et al. 2000] 2003年頃の段階で,定量性を得るために,一つの組織から100万タ グを超えるデータを取ることを目標にしていた. CAGE [Shiraki et al. 2003], 5 -SAGE [Hashimoto et al. 2004] RNA-seq [Ryan et al., 2008, Maher et al. 2009] ハイブリベースの方法 予め,各遺伝子に対応したプローブを用意しておき,そこにハイブリし たmRNAタグの量を,蛍光量などに変換して,観測する マイクロアレイ [Tse-Wen, 1983, Schena et al. 1995]
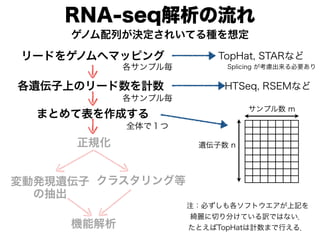
- 6. RNA-seq解析の流れ リードをゲノムへマッピング TopHat, STARなど 各遺伝子上のリード数を計数 HTSeq, RSEMなど 遺伝子数 n サンプル数 m 各サンプル毎 各サンプル毎 まとめて表を作成する 全体で1つ ゲノム配列が決定されいてる種を想定 変動発現遺伝子 の抽出 クラスタリング等 正規化 機能解析 Splicing が考慮出来る必要あり 注:必ずしも各ソフトウエアが上記を 綺麗に切り分けている訳ではない. たとえばTopHatは計数まで行える.
- 7. RNA-seq解析の流れ リードをゲノムへマッピング 各遺伝子上のリード数を計数 各サンプル毎 各サンプル毎 まとめて表を作成する 全体で1つ ゲノム配列が決定されいてる種を想定 変動発現遺伝子 の抽出 クラスタリング等 正規化 機能解析 1. RNA-seqでは,各ライブラリか ら出てくるタグ数が一致しないの で,仮想的に一致させる.遺伝子 長でもタグ数は異なる. 2. サンプルによってバイアスがあ ることがあり,必要に応じて補正 を要する. 1の例として,RPKM [Montazavi et al., 2008] 遺伝子(Exon)上の全リード * 1,000 実験で読まれた全リード(100万単位) * 遺伝子(Exon)長 2は,マイクロアレイ時にも行われていた. 非常に高発現な数遺伝子の変動に全体が ひきずられる結果,数千遺伝子が変動している ように見えてしまう.
- 8. RNA-seq解析の流れ リードをゲノムへマッピング 各遺伝子上のリード数を計数 各サンプル毎 各サンプル毎 まとめて表を作成する 全体で1つ ゲノム配列が決定されいてる種を想定 変動発現遺伝子 の抽出 クラスタリング等 正規化 機能解析 RNA-seq時代になって, Biological replicateを取るこ とが必須となっている. そのreplicateを使って,2群間比 較を行い,統計的に有意な発現変 動のある遺伝子群を抽出する edgeR [Robinson et al. 2010], DESeq [Anders et al. 2010], SAGE法の後期では,同様の研究が 行われており,その理論を RNA-seqに転用している.
- 9. 発現比率と統計的有意差 ? MA plot Robinson M D et al. Bioinformatics 2010;26:139-140 ? The Author(s) 2009. Published by Oxford University Press. (平均)発現量 発現差 Fig
- 10. 分割表による検定 ? 一般に,Fisherの正確確率検定,カイ二乗検定など. ? タグ発現解析では,ポアソン分布を用いた検定が使われる ? 二項分布を考えた場合でもpが小さい場合に相当し,ポア ソン分布で良く近似できる 150 100 1750 1900 1900 2000 Case Ctrl Total Gene1由来 Gene1以外由来 2000回のサンプルで,100回起きる事象が あるとき,1900回のサンプルで,150回事 象が起こる p=100/2000=0.05の確率で表が出るコイ ンを1900回投げ150回表が出る 単位時間tあたり,0.05t 回事象が起こると き,(150/1900)t回事象が起こる確率 二項分布 ポアソン分布
- 11. Biological replicate はどうするか ? CaseもControlも3回ずつ取られている状況を考える ? ポアソン分布の枠組みでは,Biological replicateを 直接は扱えない. ? 例えば,20回ずつ取られていれば,各遺伝子毎にt 検定も有効かもしれないが,3回では検定の検出力 が足りない ? ましてやt検定の前提条件が満たされているか,確 かめられる回数でもない. ? 実際には,統計検定が最終的な目標ではないので,「費 用の問題」「それだけ実験をするなら,他の条件を観 測したい」などで,大量のreplicate が取られることは 無い. ? とはいえ,ある程度の有意差検定を行いたい 遺伝子数n Control Case
- 12. 本当に二項分布/ポアソン分布なのだろうか? ? 二項分布の分散は np(1-p).ポアソン分布は λ(~np) ? 黒線が理論線.青点が実際の分散 ? 理論値よりも分散が遥かに大きい.特に発現量が大きい時に顕著 ? ポアソン分布で検定すると,発現量が大きい時,殆ど発現量に変 化がないのに,有意差が生まれてしまう←モデルが誤っている Anders, S., et al. (2013). Nature Protocols Fig
- 13. 負の二項分布を用いたモデル化 ? ポアソン分布に分散を表す変数を加えたい ? 負の二項分布を用いて表すモデル化が採用されている (edgeR, DESeq, cuffdiff2など) ? 負の二項分布は,ポアソン分布に変数を1つ加えたもの,あるいは, 複数のポアソン分布の混合分布として計算することが可能. ? 計測点が3点のみでは計算した分散の値が信頼出来ない問題は解消さ れていないことに注意. P(Y = y) = ? n y ◆ py (1 p)n y 二項分布: 負の二項分布: の値をとっているわけではなく, など,様々な状態を反映している 全に一致した値になることは無い NA-seq を利用した多くの 2 群間 群から複数回のサンプル(生物学 ,2 群間の比較が行われる.各遺 応の無い 2 群間比較の問題と考え で行われる実験の回数は,実験費 意の難しさから,各群の実験が 3 少ないことも多い.この少ない実 題となる. 用される検定として t 検定(ス 挙げられる.t 検定では,2 群間 来かを検定する. の RNA-seq を行ったとする.そ , Aa とする.同様に群 B から b れぞれ B1, B2, ..., Bb とする.こ ことが知られている.一方,実際にデータを調べると, 大きい所では,分散が λ より大きな値を取っている事 られている ([4] の Figure 1,あるいは [2] の Supplemn Text Figure 2.).このため,ポアソン分布を用いて検 行うと,特に発現量が大きい遺伝子に対して,本来の 上に低い P 値を算出する可能性がある. 過分散が起きた場合に,適用されるモデルが負の二 布である.負の二項分布を用いた検定は,以下のよう 式化される 定式化 4 確率変数を Y として,パラメータ p と r いると,負の二項分布は P(Y = y) = y + r ? 1 r py (1 ? p)r と表せる.また,ガンマ関数 Γ(x) = ∞ 0 e?t tx?1 dt が が自然数の時,Γ(x) = (x ? 1)! である事を用いると, P(Y = y) = Γ(y + r) Γ(r)Γ(y + 1) py (1 ? p)r となる.期待値は pr/(1 ? p),分散は pr/(1 ? p)2 であ P(Y = y) = ? y + r 1 r ◆ py (1 p)r = (y + r) (r) (y + 1) py (1 p)r 正規化の時点で離散値では なくなっているので, 連続値が扱えて嬉しい.
- 14. 変数 r を無限に飛ばすと,負の二項分布はポアソン分布に近似できる. 期待値を表す新たな変数として λ = pr 1?p を導入すると,p = λ r+λ である.こ れを,負の二項分布の式に代入して,変形する. f(y; k, r) = P(Y = y) = Γ(y + r) Γ(r)Γ(y + 1) py (1 ? p)r = λy y! · Γ(y + r) Γ(r)(r + λ)r · 1 1 + λ r r ここで r を無限に飛ばすと,第 1 項は r に依存せず,第 2 項は 1,第 3 項は 指数関数に収束するので, lim r→∞ f(y; k, r) = λy y! 1 eλ これは,期待値 λ のポアソン分布である. 証明 ■
- 15. 各遺伝子の発現量の分散を推定する ? 経験的に,分散は発現量に依存する ? 発現量が近い場合,分散も類似すると考えて 回帰問題を考えることで,分散の推定を行っ ている. ? DESeqの例:サンプルi, 遺伝子gに対し,分散 を次の式で推定する. Anders, S., et al. (2013). Nature Protocols ?(i, g) + t(i)2 ?(g) 正規化後の 推定発現量 サンプルの 総タグ数 パラメータ 遺伝子毎の値. この値を回帰で 求める 過分散を表す項 Fig
- 16. 分布は推定できた.検定はどうする? ? 分布が複雑で,解析的にはp値が求まらない. ? 求めた負の二項分布に従った乱数を発生させ,シミュレーションでp 値を求める (DESeq) ? あるいは,フィッシャーの正確確率検定の様に,観測された値以上に 極端な場合を数え上げる (edgeR) ? 例えばDESeqの戦略では ? 遺伝子g由来のタグがControl から NA回,CaseからNB回が観測された とすると ? Control と Caseは独立だと仮定するしPr(Y=NA)Pr(Y=NB)を計算 ? 負の二項分布から乱数を2個(N1, N2)発生させ Pr(Y=N1)Pr(Y=N2)を計算 ? 元の値より,p値が小さくなるような乱数の割合がp値 ? 最後は,False Discovery Rate (FDR)によって,多重検定補正を行う
- 17. Cuffdiffについて ? Cuffdiff(2)は,edgeRやDESeqと違い,RNA-seq,特にSplicing variant を定量化する話が根本にある. ? 1つの遺伝子が複数のスプライシングバリアントを保つ場合, 各リードがどのスプライシングバリアントに属するかを,最 尤推定を用いて定式化 ? 発現量の分散モデルに関しては,DESeqのモデルを踏襲 ? 但し,各exonを負の二項分布で表して,その混合分布(ベー タ負の二項分布)を発現のモデルにしている ? 最近は,edgeRやDESeqも,スプライスバリアントの定量に力 を入れているようである.
- 18. まとめと今後の課題 ? RNA-seqの導入によって,マイクロアレイに比べて定量性が高まっただ けでなく,タグをランダムサンプリングするモデルが導入でき,統計的 なモデル化が進んだ ? 現在まで,(SAGE法の延長による)過分散を考慮した発現差の統計解 析(DESeq, edgeR)と,RNA-seqから生まれた選択的スプライシング解析 (cuffdiff)の2つの道で研究が進んでおり,これらの融合が進んでいる. ? これ以上モデルを複雑にすることは,オーバーフィットとの戦いになる のではないかと思われる. ? 今後の方向性として ? アリル特異性の解析 [Akama et al. NAR 2014] ? 多サンプルに対する解析 ? クラスタ分析との融合など,が考えられるだろう. ? RNA-seqが,PacBioなどを利用した全長観測可能なものになると, スプライシングのモデル化が容易になる可能性がある.

![大規模発現量観測の小史
各遺伝子由来のタグの計数
ランダムに抽出したmRNA中の,特定の部位の配列を特定し,遺伝子発
現量の推定を行う方法
SAGE [Velculescu et al. 1995], BodyMap [Kawamoto et al. 2000]
2003年頃の段階で,定量性を得るために,一つの組織から100万タ
グを超えるデータを取ることを目標にしていた.
CAGE [Shiraki et al. 2003], 5 -SAGE [Hashimoto et al. 2004]
RNA-seq [Ryan et al., 2008, Maher et al. 2009]
ハイブリベースの方法
予め,各遺伝子に対応したプローブを用意しておき,そこにハイブリし
たmRNAタグの量を,蛍光量などに変換して,観測する
マイクロアレイ [Tse-Wen, 1983, Schena et al. 1995]](https://image.slidesharecdn.com/20141217sigbiopub-141217082915-conversion-gate01/85/RNAseq-A-Review-3-320.jpg)
![RNA-seq解析の流れ
リードをゲノムへマッピング
各遺伝子上のリード数を計数
各サンプル毎
各サンプル毎
まとめて表を作成する
全体で1つ
ゲノム配列が決定されいてる種を想定
変動発現遺伝子
の抽出
クラスタリング等
正規化
機能解析
1. RNA-seqでは,各ライブラリか
ら出てくるタグ数が一致しないの
で,仮想的に一致させる.遺伝子
長でもタグ数は異なる.
2. サンプルによってバイアスがあ
ることがあり,必要に応じて補正
を要する.
1の例として,RPKM [Montazavi et al., 2008]
遺伝子(Exon)上の全リード * 1,000
実験で読まれた全リード(100万単位) * 遺伝子(Exon)長
2は,マイクロアレイ時にも行われていた.
非常に高発現な数遺伝子の変動に全体が
ひきずられる結果,数千遺伝子が変動している
ように見えてしまう.](https://image.slidesharecdn.com/20141217sigbiopub-141217082915-conversion-gate01/85/RNAseq-A-Review-7-320.jpg)
![RNA-seq解析の流れ
リードをゲノムへマッピング
各遺伝子上のリード数を計数
各サンプル毎
各サンプル毎
まとめて表を作成する
全体で1つ
ゲノム配列が決定されいてる種を想定
変動発現遺伝子
の抽出
クラスタリング等
正規化
機能解析
RNA-seq時代になって,
Biological replicateを取るこ
とが必須となっている.
そのreplicateを使って,2群間比
較を行い,統計的に有意な発現変
動のある遺伝子群を抽出する
edgeR [Robinson et al. 2010],
DESeq [Anders et al. 2010],
SAGE法の後期では,同様の研究が
行われており,その理論を
RNA-seqに転用している.](https://image.slidesharecdn.com/20141217sigbiopub-141217082915-conversion-gate01/85/RNAseq-A-Review-8-320.jpg)




![負の二項分布を用いたモデル化
? ポアソン分布に分散を表す変数を加えたい
? 負の二項分布を用いて表すモデル化が採用されている (edgeR,
DESeq, cuffdiff2など)
? 負の二項分布は,ポアソン分布に変数を1つ加えたもの,あるいは,
複数のポアソン分布の混合分布として計算することが可能.
? 計測点が3点のみでは計算した分散の値が信頼出来ない問題は解消さ
れていないことに注意.
P(Y = y) =
?
n
y
◆
py
(1 p)n y
二項分布:
負の二項分布:
の値をとっているわけではなく,
など,様々な状態を反映している
全に一致した値になることは無い
NA-seq を利用した多くの 2 群間
群から複数回のサンプル(生物学
,2 群間の比較が行われる.各遺
応の無い 2 群間比較の問題と考え
で行われる実験の回数は,実験費
意の難しさから,各群の実験が 3
少ないことも多い.この少ない実
題となる.
用される検定として t 検定(ス
挙げられる.t 検定では,2 群間
来かを検定する.
の RNA-seq を行ったとする.そ
, Aa とする.同様に群 B から b
れぞれ B1, B2, ..., Bb とする.こ
ことが知られている.一方,実際にデータを調べると,
大きい所では,分散が λ より大きな値を取っている事
られている ([4] の Figure 1,あるいは [2] の Supplemn
Text Figure 2.).このため,ポアソン分布を用いて検
行うと,特に発現量が大きい遺伝子に対して,本来の
上に低い P 値を算出する可能性がある.
過分散が起きた場合に,適用されるモデルが負の二
布である.負の二項分布を用いた検定は,以下のよう
式化される
定式化 4 確率変数を Y として,パラメータ p と r
いると,負の二項分布は
P(Y = y) =
y + r ? 1
r
py
(1 ? p)r
と表せる.また,ガンマ関数 Γ(x) =
∞
0
e?t
tx?1
dt が
が自然数の時,Γ(x) = (x ? 1)! である事を用いると,
P(Y = y) =
Γ(y + r)
Γ(r)Γ(y + 1)
py
(1 ? p)r
となる.期待値は pr/(1 ? p),分散は pr/(1 ? p)2
であ
P(Y = y) =
?
y + r 1
r
◆
py
(1 p)r
=
(y + r)
(r) (y + 1)
py
(1 p)r
正規化の時点で離散値では
なくなっているので,
連続値が扱えて嬉しい.](https://image.slidesharecdn.com/20141217sigbiopub-141217082915-conversion-gate01/85/RNAseq-A-Review-13-320.jpg)




![まとめと今後の課題
? RNA-seqの導入によって,マイクロアレイに比べて定量性が高まっただ
けでなく,タグをランダムサンプリングするモデルが導入でき,統計的
なモデル化が進んだ
? 現在まで,(SAGE法の延長による)過分散を考慮した発現差の統計解
析(DESeq, edgeR)と,RNA-seqから生まれた選択的スプライシング解析
(cuffdiff)の2つの道で研究が進んでおり,これらの融合が進んでいる.
? これ以上モデルを複雑にすることは,オーバーフィットとの戦いになる
のではないかと思われる.
? 今後の方向性として
? アリル特異性の解析 [Akama et al. NAR 2014]
? 多サンプルに対する解析
? クラスタ分析との融合など,が考えられるだろう.
? RNA-seqが,PacBioなどを利用した全長観測可能なものになると,
スプライシングのモデル化が容易になる可能性がある.](https://image.slidesharecdn.com/20141217sigbiopub-141217082915-conversion-gate01/85/RNAseq-A-Review-18-320.jpg)