Visualizing biological graphs in Cytoscape.js
- 1. G R A P H S I N C Y T O S C A P E . J S V I S U A L I Z I N G B I O L O G I C A L Ben Keller? linkedin.com/in/bjkeller Seattle/Bellevue Graphs - Data, Processing, Algorithms Meetup? 6 May 2014
- 2. B I O L O G I C A L G R A P H S
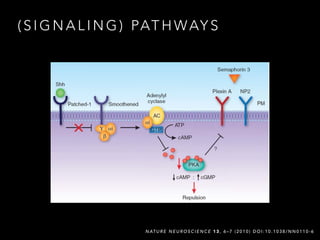
- 3. ( S I G N A L I N G ) PAT H WAY S N AT U R E N E U R O S C I E N C E 1 3 , 6 ©C 7 ( 2 0 1 0 ) D O I : 1 0 . 1 0 3 8 / N N 0 1 1 0 - 6
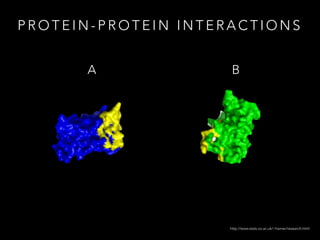
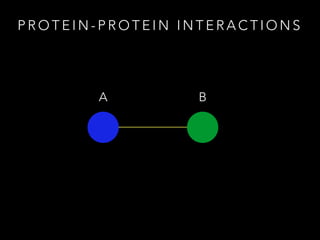
- 4. P R O T E I N - P R O T E I N I N T E R A C T I O N S A B http://www.stats.ox.ac.uk/~hamer/research.html
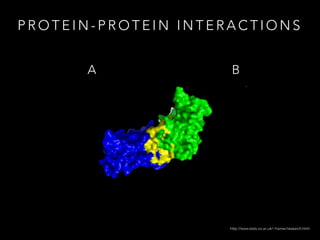
- 5. P R O T E I N - P R O T E I N I N T E R A C T I O N S A B http://www.stats.ox.ac.uk/~hamer/research.html
- 6. P R O T E I N - P R O T E I N I N T E R A C T I O N S A B
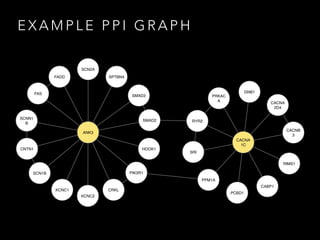
- 7. E X A M P L E P P I G R A P H ANK3 CABP1 CACNA 2D4 CACNB 3 GNB1 PCBD1 RIMS1 PPM1A PRKAC A RYR2 SRI CACNA 1C FAS FADD CNTN1 CRKL HOOK1 KCNC1 KCNC2 SMAD3 SMAD2 PIK3R1 SPTBN4 SCN1B SCNN1 B SCN2A
- 8. AT T R I B U T E S Tell us something about gene/protein: ? Where it occurs ? What it does ? What processes it contributes to ? What diseases it is implicated in Allow us to ask ? do these genes belong together? ? do they work together?
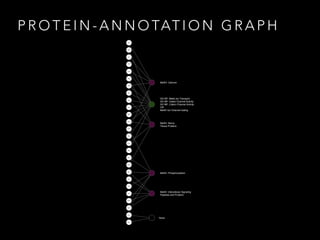
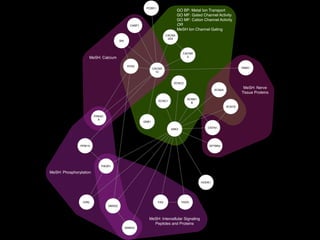
- 9. P R O T E I N - A N N O TAT I O N G R A P H MeSH: Calcium GO BP: Metal Ion Transport GO MF: Gated Channel Activity GO MF: Cation Channel Activity OR MeSH Ion Channel Gating MeSH: Phosphorylation MeSH: Intercellular Signaling Peptides and Proteins MeSH: Nerve Tissue Proteins CRKL ANK3 CABP1 CACNA 2D4 CACNB 3 GNB1 PCBD1 RIMS1 PPM1A PRKAC A RYR2 SRI CACNA 1C FAS FADD CNTN1 HOOK1 KCNC1 KCNC2 SMAD3 SMAD2 PIK3R1 SPTBN4 SCN1B SCNN1 B SCN2A None
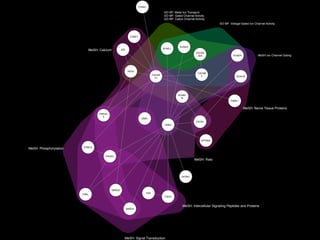
- 10. C O N C E P T G R A P H MeSH: Calcium GO BP: Metal Ion Transport GO MF: Gated Channel Activity GO MF: Cation Channel Activity OR MeSH Ion Channel Gating MeSH: Phosphorylation MeSH: Intercellular Signaling Peptides and Proteins MeSH: Nerve Tissue Proteins CRKL ANK3 CABP1 CACNA 2D4 CACNB 3 GNB1 PCBD1 RIMS1 PPM1A PRKAC A RYR2 SRI CACNA 1C FAS FADD CNTN1 HOOK1 KCNC1 KCNC2 SMAD3 SMAD2 PIK3R1 SPTBN4 SCN1B SCNN1 B SCN2A None MeSH: Calcium GO BP: Metal Ion Transport GO MF: Gated Channel Activity GO MF: Cation Channel Activity OR MeSH Ion Channel Gating MeSH: Phosphorylation MeSH: Intercellular Signaling Peptides and Proteins MeSH: Nerve Tissue Proteins
- 11. MeSH: Calcium GO BP: Metal Ion Transport GO MF: Gated Channel Activity GO MF: Cation Channel Activity GO MF: Voltage-Gated Ion Channel Activity MeSH: Phosphorylation MeSH: Signal Transduction MeSH: Intercellular Signaling Peptides and Proteins MeSH: Rats MeSH: Nerve Tissue Proteins MeSH Ion Channel Gating CRKL ANK3 CABP1 CACNA 2D4 CACNB 3 GNB1 PCBD1 RIMS1 PPM1A PRKAC A RYR2 SRI CACNA 1C FAS FADD CNTN1 HOOK1 KCNC1 KCNC2 SMAD3 SMAD2 PIK3R1 SPTBN4 SCN1B SCNN1 B SCN2A
- 12. MeSH: Calcium GO BP: Metal Ion Transport GO MF: Gated Channel Activity GO MF: Cation Channel Activity OR MeSH Ion Channel Gating MeSH: Phosphorylation MeSH: Intercellular Signaling Peptides and Proteins MeSH: Nerve Tissue Proteins CRKL ANK3 CABP1 CACNA 2D4 CACNB 3 GNB1 PCBD1 RIMS1 PPM1A PRKAC A RYR2 SRI CACNA 1C FAS FADD CNTN1 HOOK1 KCNC1 KCNC2 SMAD3 SMAD2 PIK3R1 SPTBN4 SCN1B SCNN1 B SCN2A
- 13. C Y T O S C A P E . J S
- 15. ( J S B I N ) H T M L <!DOCTYPE html> <html> <head> <meta charset="utf-8"> <title>Meetup Cytoscape.js Lab</title> <script src=/slideshow/cytoscapejs0514/34499506/"http:/ajax.googleapis.com/ajax/libs/jquery/1/jquery.min.js"></script> <script src="http://cytoscape.github.io/cytoscape.js/api/cytoscape.js-latest/cytoscape.min.js"></script> </head> <body> <div id="cy" style="height:90%;width:90%;position:absolute;left:0;top:0;"></div> </body> </html> ? jQuery ? cytoscape.js ? (your script) container for graph
- 16. J S F I L E $(document).ready(function(){ //cytoscape code goes here });
- 17. C Y T O S C A P E S E S S I O N var cy = cytoscape({ container: document.getElementById('cy'), ready: function() {} });
- 18. G R A P H var graphelements = { nodes: [ { data: { id: 'n1', name: Ī«A' } }, { data: { id: 'n2', name: Ī«B' } }, { data: { id: 'n3', name: Ī«C' } }, { data: { id: 'n4', name: Ī«D' } } ], edges: [ { data: { id: 'e1', source: 'n1', target: 'n2' } }, { data: { id: 'e2', source: 'n3', target: 'n2' } }, { data: { id: 'e3', source: 'n4', target: 'n2' } } ] };
- 20. A D D : S H O W N O D E L A B E L O N H O V E R cy.on('mouseover','node',function(evt) { var node = evt.cyTarget; node.css('content','data(name)'); }); cy.on('mouseout','node',function(evt){ var node = evt.cyTarget; node.css('*',''); }); (add after session definition)http://jsbin.com/reqeg/12/edit
- 21. A D D : S E T U P F O R S T Y L E S var nodestyle = {}; var edgestyle = {}; ! var stylesheet = cytoscape.stylesheet().? selector("node").css(nodestyle). selector(Ī░edge").css(edgestyle); ! var cy = cytoscape({ container: document.getElementById('cy'), style: stylesheet, ready: function() {} }); http://jsbin.com/reqeg/11/edit
- 22. A D D : S E T N O D E S T Y L E var nodestyle = { 'font-size': '12pt', 'min-zoomed-font-size': Ī«9pt', 'text-halign': 'center', 'text-valign': 'center', 'color': 'white' }; http://jsbin.com/reqeg/10/edit
- 23. A D D : M A P P I N G AT T R I B U T E S ( 1 ) var nodestyle = { 'font-size': '12pt', 'min-zoomed-font-size': '9pt', 'text-valign': 'center', 'text-halign': 'center', 'color': 'white', 'width': 'mapData(degree,0,5,20,80)', 'height': 'mapData(degree,0,5,20,80)' }; http://jsbin.com/reqeg/9/edit
- 24. A D D : M A P P I N G AT T R I B U T E S ( 2 ) var degmap = {}; var nodes = cy.nodes(); for (var i = 0; i < nodes.length; i++) { degmap[nodes[i].id()] = { degree: nodes[i].degree() }; } cy.batchData(degmap); http://jsbin.com/reqeg/7/edit
- 26. L AY O U T S var layoutopts = { name: 'breadthfirst', roots: ['n1','n26'] }; cy.layout(layoutopts); ? random ? preset ? grid ? circle ? concentric ? breadthfirst ? arbor * ? cose ? null (for algorithms)
- 27. A R B O R ( F O R C E - D I R E C T E D ) <script src=/slideshow/cytoscapejs0514/34499506/"../node_modules/cytoscape/lib/arbor.js"></script> <script src="../node_modules/cytoscape/dist/cytoscape.min.js"></script>









![G R A P H
var graphelements = {
nodes: [
{ data: { id: 'n1', name: Ī«A' } },
{ data: { id: 'n2', name: Ī«B' } },
{ data: { id: 'n3', name: Ī«C' } },
{ data: { id: 'n4', name: Ī«D' } }
],
edges: [
{ data: { id: 'e1', source: 'n1', target: 'n2' } },
{ data: { id: 'e2', source: 'n3', target: 'n2' } },
{ data: { id: 'e3', source: 'n4', target: 'n2' } }
]
};](https://image.slidesharecdn.com/cytoscapejs0514-140509184213-phpapp01/85/Visualizing-biological-graphs-in-Cytoscape-js-18-320.jpg)





![A D D : M A P P I N G AT T R I B U T E S ( 2 )
var degmap = {};
var nodes = cy.nodes();
for (var i = 0; i < nodes.length; i++) {
degmap[nodes[i].id()] = { degree: nodes[i].degree() };
}
cy.batchData(degmap);
http://jsbin.com/reqeg/7/edit](https://image.slidesharecdn.com/cytoscapejs0514-140509184213-phpapp01/85/Visualizing-biological-graphs-in-Cytoscape-js-24-320.jpg)

![L AY O U T S
var layoutopts = {
name: 'breadthfirst',
roots: ['n1','n26']
};
cy.layout(layoutopts);
? random
? preset
? grid
? circle
? concentric
? breadthfirst
? arbor *
? cose
? null (for algorithms)](https://image.slidesharecdn.com/cytoscapejs0514-140509184213-phpapp01/85/Visualizing-biological-graphs-in-Cytoscape-js-26-320.jpg)

