ECE 565 Project1
0 likes411 views
This document contains code and explanations for image processing tasks involving edge detection, thresholding, and segmentation. It includes: 1) Code to perform spatial filtering of an image using a 3x3 mask and Sobel edge detection filters. 2) Application of the code to detect edges and segment a large blood vessel in a kidney image. 3) Implementation of Otsu's thresholding algorithm to automatically select a threshold for segmentation. 4) Comparison of Otsu's method to global thresholding on a polymersomes image, finding Otsu's performs better.
1 of 11
Downloaded 18 times
![% we have a1 to a9 as variables
w =[a1 a2 a3;a4 a5 a6;a7 a8 a9] % w is the filter (assumed to be 3x3)
g=zeros(x+2,y+2);% The original image is padded with 0's
for i=1:x
for j=1:y
g(i+1,j+1)=f(i,j);
end
end
%cycle through the array and apply the filter
for i=1:x
for j=1:y
img(i,j)=g(i,j)*w(1,1)+g(i+1,j)*w(2,1)+g(i+2,j)*w(3,1) ... %first column
+ g(i,j+1)*w(1,2)+g(i+1,j+1)*w(2,2)+g(i+2,j+1)*w(3,2)... %second column
+ g(i,j+2)*w(1,3)+g(i+1,j+2)*w(2,3)+g(i+2,j+2)*w(3,3);
end
end
figure
imshow(img,[])
(b).
f=imread('..');
figure
imshow(f); %original figure
w =[a1 a2 a3;a4 a5 a6;a7 a8 a9] %filter from (a)
c = conv2(f,w,'same'); %image after convolution with mask
s1 = [-1 -2 -1;0 0 0;1 2 1]; % sobel operator in horizontal
s2 = [-1 0 1;-2 0 2;-1 0 1]; % sobel operator in vertical
gx = conv2(c,s1,'same');
gy = conv2(c,s2,'same');
M = sqrt(gx.*gx+gy.*gy); % maginitude
figure % image after convolution with horizontal sobel operator
imshow(gx);
figure % image after convolution with vertical sobel operator
imshow(gy)
g = gx+gy; % image with horizontal gx plus vertical gy
figure](https://image.slidesharecdn.com/ba4b1a67-e0dc-44f9-bef4-291be5a9cc58-160523230201/85/ECE-565-Project1-2-320.jpg)
![imshow(g)
% historgram stem of image
I = gpuArray(imread('E:myimagesÂĨkidney.tif'));
[counts,x] = imhist(I);
stem(x,counts);
BW = im2bw(f,T);
figure
imshow(BW)
(c).
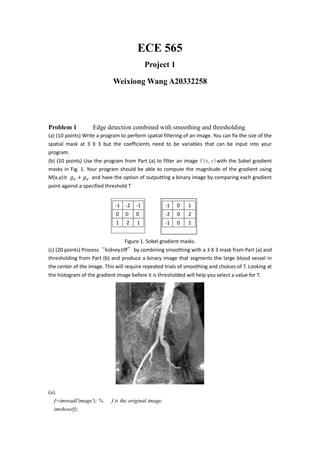
Figure before smoothing Figure after smoothing
To produce a binary image thatsegments the large blood vessel in the center of the image.](https://image.slidesharecdn.com/ba4b1a67-e0dc-44f9-bef4-291be5a9cc58-160523230201/85/ECE-565-Project1-3-320.jpg)
![Code:
f=imread('E:myimagesÂĨkidney.tif');
figure
imshow(f); %original figure
w=ones(3)/9; %filter from (a)
c = conv2(f,w,'same'); %image after convolution with mask
s1 = [-1 -2 -1;0 0 0;1 2 1]; % sobel operator in horizontal
s2 = [-1 0 1;-2 0 2;-1 0 1]; % sobel operator in vertical
gx = conv2(c,s1,'same');
gy = conv2(c,s2,'same');
M = sqrt(gx.*gx+gy.*gy); % maginitude
figure % image after convolution with horizontal sobel operator
imshow(gx);
figure % image after convolution with vertical sobel operator
imshow(gy)
g = gx+gy; % image with horizontal gx plus vertical gy
figure
imshow(g)
%historgram stem of image
[counts,x] = imhist(f);
stem(x,counts);
title('Stem of kindney.tif')
xlabel('Intensity level')
ylabel('Number of pixels')
axis([0 256 0 20000])
figure;
imhist(f,64)
title('Histogram of kindney.tif')
xlabel('Intensity level')
ylabel('Number of pixels')
BW = im2bw(f,0.72);
figure
imshow(BW)](https://image.slidesharecdn.com/ba4b1a67-e0dc-44f9-bef4-291be5a9cc58-160523230201/85/ECE-565-Project1-6-320.jpg)

![function level = g_t(I)
I = imread('E:myimagesÂĨnoisy_fingerprint.tif');
% STEP 1: Compute mean intensity of image from histogram, set T=mean(I)
[counts,N]=imhist(I);
i=1;
mu=cumsum(counts);
T(i)=(sum(N.*counts))/mu(end);
T(i)=round(T(i));
% STEP 2: compute Mean above T (MAT) and Mean below T (MBT) using T from
% step 1
mu2=cumsum(counts(1:T(i)));
MBT=sum(N(1:T(i)).*counts(1:T(i)))/mu2(end);
mu3=cumsum(counts(T(i):end));
MAT=sum(N(T(i):end).*counts(T(i):end))/mu3(end);
i=i+1;
% new T = (MAT+MBT)/2
T(i)=round((MAT+MBT)/2);
% STEP 3 to n: repeat step 2 if T(i)~=T(i-1)
while abs(T(i)-T(i-1))>=1
mu2=cumsum(counts(1:T(i)));
MBT=sum(N(1:T(i)).*counts(1:T(i)))/mu2(end);
mu3=cumsum(counts(T(i):end));
MAT=sum(N(T(i):end).*counts(T(i):end))/mu3(end);
i=i+1;
T(i)=round((MAT+MBT)/2);
Threshold=T(i);
end
% Normalize the threshold to the range [i, 1].
level = (Threshold - 1) / (N(end) - 1);
BW = im2bw(I,level);
imshow(BW)](https://image.slidesharecdn.com/ba4b1a67-e0dc-44f9-bef4-291be5a9cc58-160523230201/85/ECE-565-Project1-8-320.jpg)
![Compare the result for a) and b)
We see the Otsu has a better performance than global,because Otsu is based on the histogram,so
we can get a good threshold value than in global thresholding.In Otsu,We canât change the
distributions, but we can adjust where we separate them (the threshold). As we adjust the threshold
one way, we increase the spread of one and decrease the spread of the other.
Code:
I = imread('E:myimagesÂĨpolymersomes.tif');
mgk = 0;
mt = 0;
[m,n] = size(I);
h = imhist(I);
pi = h/(m.*n);
for i=1:1:256
if pi(i)~=0
lv=i;
break
end
end
for i=256:-1:1
if pi(i)~=0
hv=i;
break
end
end
lh = hv - lv;
for k = 1:256
p1(k)=sum(pi(1:k));
p2(k)=sum(pi(k+1:256));
end
for k=1:256
m1(k)=sum((k-1)*pi(1:k))/p1(k);
m2(k)=sum((k-1)*pi(k+1:256))/p2(k);
end
for k=1:256
mgk=(k-1)*pi(k)+mgk;
end
for k =1:256
var(k)=p1(k)*(m1(k)-mgk)^2+p2(k)*(m2(k)-mgk)^2;
end
[y,T]=max(var(:));
T=T+lv;](https://image.slidesharecdn.com/ba4b1a67-e0dc-44f9-bef4-291be5a9cc58-160523230201/85/ECE-565-Project1-10-320.jpg)

Recommended
Project2



Project2?? ?
Ėý
The document describes a project involving image processing and boundary analysis. It includes:
1. Smoothing an image, thresholding it to binary, extracting the outer boundary, subsampling the boundary, and computing the Freeman chain code and descriptors of the subsampled boundary.
2. Extracting the boundary of an image of a chromosome, computing the Fourier descriptors of the boundary, and reconstructing the boundary using different percentages of the descriptors.
3. The provided code computes Freeman chain codes, Fourier descriptors, and performs other boundary analysis tasks like subsampling and reconstruction from descriptors.Computer graphics 2



Computer graphics 2Prabin Gautam
Ėý
The document discusses computer graphics concepts like points, pixels, lines, and circles. It begins with definitions of pixels and how they relate to points in geometry. It then covers the basic structure for specifying points in OpenGL and how to draw points, lines, and triangles. Next, it discusses algorithms for drawing lines, including the digital differential analyzer (DDA) method and Bresenham's line algorithm. Finally, it covers circle drawing and introduces the mid-point circle algorithm. In summary:
1) It defines key computer graphics concepts like pixels, points, lines, and circles.
2) It explains the basic OpenGL functions for drawing points and lines and provides examples of drawing simple shapes.
3) ItCgm Lab Manual



Cgm Lab ManualOriental College of Technology,Bhopal
Ėý
This document is a lab manual for a Computer Graphics and Multimedia course. It includes:
- An introduction and contact information for the instructor.
- A table of contents listing various sections such as the course timetable, syllabus, list of programs to be completed, and important exam questions.
- Details of the course syllabus which covers topics like raster graphics, 2D and 3D transformations, color models, multimedia systems, compression techniques and more.
- A list of 11 programming experiments related to computer graphics algorithms and multimedia, including DDA line drawing, Bresenham's circle algorithm, transformations and compression.
- Descriptions and code for the first 3 experiments - DDA lineIntro to scan conversion



Intro to scan conversionMohd Arif
Ėý
Scan conversion is the process of representing continuous graphics objects as discrete pixels. It involves converting geometric primitives like lines and circles, defined by parameters, into a set of pixels that make up the primitive in an image. This involves mapping real-valued coordinates to integer pixel coordinates. One approach is to take the floor of the x and y values, while another is to take the floor of x+0.5 and y+0.5 to center the coordinate system at the pixel grid.Line Drawing Algorithms - Computer Graphics - Notes



Line Drawing Algorithms - Computer Graphics - NotesOmprakash Chauhan
Ėý
Straight-line drawing algorithms are based on incremental methods.
In incremental method line starts with a straight point, then some fix incrementable is added to current point to get next point on the line and the same has continued all the end of the line.
Test



TestKinni MEW
Ėý
This document contains solutions to two questions about image processing techniques. Question 1 involves implementing Sobel edge detection using 4 convolution masks and calculating the approximate gradient magnitude. Question 2 involves implementing a corner detector by building a neighborhood matrix around each pixel, computing its singular values via SVD, and highlighting locations where the second singular value exceeds a threshold after removing neighboring duplicates.Line drawing algorithm and antialiasing techniques



Line drawing algorithm and antialiasing techniquesAnkit Garg
Ėý
The document discusses computer graphics and line drawing algorithms. Module 1 covers introduction to graphics hardware, display devices, and graphics software. Module 2 discusses output primitives like lines, circles, ellipses, and clipping algorithms like Cohen-Sutherland and Sutherland-Hodgeman. It then explains the Digital Differential Algorithm (DDA) and Bresenham's line drawing algorithms for scan converting lines. DDA calculates increments in the x or y direction based on the slope. Bresenham's uses only integer calculations. Both algorithms are demonstrated with examples. The document also discusses anti-aliasing techniques like supersampling and area sampling to reduce jagged edges.Computer graphics lab report with code in cpp



Computer graphics lab report with code in cppAlamgir Hossain
Ėý
This is the lab report for computer graphics in cpp language. Basically this course is only for the computer science and engineering students.
Problem list:
1.Program for the generation of Bresenham Line Drawing.
2. Program for the generation of Digital Differential Analyzer (DDA) Line Drawing.
3. Program for the generation of Midpoint Circle Drawing.
4. Program for the generation of Midpoint Ellipse Drawing.
5. Program for the generation of Translating an object.
6. Program for the generation of Rotating an Object.
7. Program for the generation of scaling an object.
All programs are coaded in cpp language .Computer graphics 



Computer graphics shafiq sangi
Ėý
This document contains information about computer graphics including:
1. An introduction to computer graphics, its applications, and basic functions like drawing lines, circles, rectangles.
2. Details of 4 assignments - the first involves studying graphics functions, the second develops a CPU architecture GUI, the third defines a point class, and the fourth defines line and circle drawing classes.
3. The last assignment involves writing C++ classes to draw lines using DDA and Bresenham's algorithms and inheriting from the point class, and a circle class inheriting from the line class.Computer graphics lab manual



Computer graphics lab manualUma mohan
Ėý
The document provides source code for generating and manipulating computer graphics using various algorithms. It includes algorithms for drawing lines, circles and curves, as well as algorithms for translating, rotating, and scaling two-dimensional and three-dimensional objects. The source code is written in C/C++ and uses graphics libraries to output the results. Various input parameters are taken from the user and output is displayed to demonstrate the algorithms.Computer graphics 



Computer graphics Prianka Padmanaban
Ėý
The document describes algorithms for 2D transformations in computer graphics using C programming language. It discusses functions for translation, rotation, and scaling of 2D objects.
The algorithms take input for the type of transformation, parameters for the transformation, and coordinates of the 2D object. Translation simply adds the translation offsets to the x and y coordinates. Rotation rotates the object around the x-axis or y-axis based on the angle input. Scaling multiplies the x and y coordinates by respective scaling factors.
The transformed object is displayed on the graphics screen. Thus basic 2D transformations like translation, rotation, scaling are implemented through these algorithms in C.Basics of Computer graphics lab



Basics of Computer graphics labPriya Goyal
Ėý
This document provides instructions for drawing basic shapes in graphics mode in Turbo C++. It explains how to enable the graphics library, initialize graphics mode, and use functions like putpixel, line, rectangle, circle, ellipse, arc, bar and bar3d to draw pixels, lines, rectangles, circles, ellipses, arcs, filled rectangles and 3D filled rectangles. Examples of code are given to draw each shape. Programming assignments are provided to combine the shapes to draw a flag, hut, smiley face and fish.Computer graphics lab manual



Computer graphics lab manualAnkit Kumar
Ėý
The document provides a lab manual for computer graphics experiments in C language. It includes experiments on digital differential analyzer algorithm, Bresenham's line drawing algorithm, midpoint circle generation algorithm, ellipse generation algorithm, text and shape creation, 2D and 3D transformations, curve generation, and basic animations. It outlines the hardware and software requirements to run the experiments and provides background, algorithms, sample programs and outputs for each experiment.Computer graphics



Computer graphicsNanhen Verma
Ėý
The document discusses algorithms for drawing circles and filling polygons on a computer screen. It covers the mid-point circle algorithm for determining pixel positions on a circle, as well as boundary filling and flood filling algorithms for coloring the interior of polygon shapes. The mid-point circle algorithm uses a decision parameter to iteratively calculate pixel coordinates on the circle path. Filling algorithms like boundary fill use recursion to color neighboring pixels of the same color as the initially selected point.Cg lab cse-v (1) (1)



Cg lab cse-v (1) (1)Surya Sukumaran
Ėý
The document contains an algorithm for drawing an ellipse using the midpoint algorithm. It initializes the graphics system and gets user input for the major and minor radii. It calculates an initial decision parameter P and plots the first point. It then loops while a condition on the x and y coordinates is met, incrementing x by 1 each iteration. The decision parameter P is updated each iteration based on whether it is greater than or less than 0. Symmetric points are plotted in each quadrant to draw the ellipse. After drawing across the minor axis, it continues the algorithm to draw across the major axis by initializing new parameters and looping while y is not 0, updating the decision parameter P1 and decrementing y each iteration.Dda algo notes



Dda algo notesshreeja asopa
Ėý
The document discusses three algorithms for line generation in computer graphics:
1. The DDA algorithm calculates increments in x and y coordinates between two endpoints and plots pixels along the line incrementally.
2. Bresenham's algorithm uses integer calculations and chooses pixel coordinates closest to the true line by comparing differences above and below the line.
3. The mid-point algorithm calculates the midpoint between potential next points and chooses the one closest to the true line based on whether the intersection is above or below the midpoint.Cgm Lab Manual



Cgm Lab ManualOriental College of Technology,Bhopal
Ėý
The document is a lab manual for a course on Computer Graphics and Multimedia. It contains:
1. A table of contents listing various sections like the time table, university scheme, syllabus, list of books, and list of programs.
2. The time table, university scheme, and syllabus provide details about the course schedule, assessment scheme, and topics to be covered.
3. The list of books and list of programs provide resources for students to refer to for the course and experiments to be performed in the lab.Computer graphics lab assignment



Computer graphics lab assignmentAbdullah Al Shiam
Ėý
The document contains 10 programs written in C programming language to perform various 2D and 3D graphics operations like drawing lines, rectangles, circles, ellipses, torus, scaling, rotating, shearing and reflecting objects. Program 1-5 demonstrate drawing basic shapes like line, rectangle, circle and ellipse. Program 6 draws a 3D torus using OpenGL. Program 7-9 demonstrate transformations like scaling, rotating and shearing of objects. Program 10 shows reflection of an object about x-axis, y-axis and origin.Introduction to Data Science With R Lab Record



Introduction to Data Science With R Lab RecordLakshmi Sarvani Videla
Ėý
This document provides an index and overview of programs related to data science concepts in R. The programs cover topics like arithmetic operations on vectors, matrix operations, graphs, loops, and functions. The index lists 8 programs from August to October 2021 covering these topics. For each program, there is a brief description of the concepts covered and examples of R code and output.Matlab plotting



Matlab plottingAmr Rashed
Ėý
The document contains 20 MATLAB programs demonstrating various plotting and data visualization techniques including:
- Plotting sinusoidal waves and applying half/full wave rectification
- Converting between polar and Cartesian coordinates and plotting circles
- Generating and plotting cylinder surfaces
- Using EZ functions like ezplot, ezsurf, and ezpolar to plot functions
- Taking derivatives and plotting functions and their derivatives
- Plotting noise signals and calculating rate of change over time
- Generating 2D and 3D surfaces from gridded data
- Plotting polar plots and converting to Cartesian coordinates
- Applying FFT and filtering to decompose a signal into frequency components
- Using pie charts and 3D pie plots toLec02 03 rasterization



Lec02 03 rasterizationMaaz Rizwan
Ėý
The document discusses different algorithms for rasterizing lines in computer graphics. It describes how rasterization works by converting vector graphics into pixel representations. It then explains three strategies for rasterizing a line between two points: using the explicit line equation, parametric form, and incremental algorithms like the Digital Differential Analyzer (DDA) algorithm. The DDA algorithm works by incrementally calculating the next x and y pixel coordinates along the line using step sizes, avoiding expensive floating-point calculations.Programming Assignment Help



Programming Assignment HelpProgramming Homework Help
Ėý
I am Gill H. I am a Programming Assignment Expert at programminghomeworkhelp.com. I hold a Ph.D. in Electronics Engineering from, the University of Texas, USA. I have been helping students with their homework for the past 8 years. I solve assignments related to Programming.
Visit programminghomeworkhelp.com or email support@programminghomeworkhelp.com. You can also call on +1 678 648 4277 for any assistance with Programming Assignments.
Matlab plotting



Matlab plottingshahid sultan
Ėý
This document discusses various MATLAB plotting commands and techniques. It begins by explaining how to create basic 2D plots using the plot() command to plot x-y data or a function. It also covers formatting plots by adding titles, labels, legends etc. The document then discusses plotting multiple graphs in the same figure using either the plot() command or hold on/off commands. Finally, it demonstrates how to create multiple subplots on one figure using the subplot command.Writeup advanced lane_lines_project



Writeup advanced lane_lines_projectManish Jauhari
Ėý
The document describes an Advanced Lane Finding project that uses computer vision techniques to identify lane boundaries in video from a front-facing car camera. The steps include: 1) removing distortion, 2) isolating lanes using color and gradient thresholds, 3) warping the image to a bird's-eye view, 4) fitting curves to the lane pixels, and 5) creating the final output image with lane and radius of curvature information overlaid. Improvement areas include automating parameter selection using machine learning rather than manual tuning.Line drawing algo.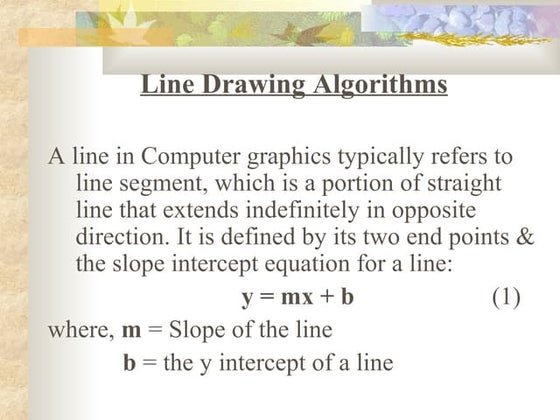



Line drawing algo.Mohd Arif
Ėý
The document discusses line drawing algorithms in computer graphics. It defines a line segment and provides equations to determine the slope and y-intercept of a line given two endpoints. It then introduces the Digital Differential Analyzer (DDA) algorithm, an incremental scan conversion method that calculates the next point on the line based on the previous point's coordinates and the line's slope. The algorithm involves less floating point computation than directly using the line equation at each step. An example demonstrates applying DDA to scan convert a line between two points. Limitations of DDA include the processing costs of rounding and floating point arithmetic as well as accumulated round-off error over long line segments.Introduction to graphics programming in c



Introduction to graphics programming in cbaabtra.com - No. 1 supplier of quality freshers
Ėý
This document provides an introduction to graphics programming in C. It discusses setting up graphics using GCC, basic concepts of graphics programming in C, common graphics functions like line(), circle(), rectangle(), and text functions like outtext() and outtextxy(). It also includes a short example program to demonstrate drawing various shapes and text.Introduction to curve



Introduction to curveOmprakash Chauhan
Ėý
In computer graphics, we often need to draw different types of objects onto the screen. Objects are not flat all the time and we need to draw curves many times to draw an object.Fuzzy dm



Fuzzy dmAkshay Chaudhari
Ėý
This document discusses fuzzy clustering, which allows data points to belong to multiple clusters with a degree of membership. It describes fuzzy c-means clustering, which was improved in 1981 by Bezdek. The algorithm involves initializing membership values, computing centroids, recomputing membership values iteratively until convergence. Pros include a more natural representation of overlapping clusters, while cons include sensitivity to initialization and needing to specify the number of clusters. Applications include image segmentation, enhancement, and change detection.3. ÐŅŅŅÐļКÐļ ÐēÐĩŅÐĩÐŧÐļКÐļ



3. ÐŅŅŅÐļКÐļ ÐēÐĩŅÐĩÐŧÐļКÐļÐĄÐĩŅÐģŅÐđ ÐÐŧŅŅÐļŅÐļÐ―
Ėý
ÐÐēÐ°Ð―ÐūÐēŅŅКа ÐĄÐēŅŅÐŧÐ°Ð―Ð° ÐÐūÐŧÐūÐīÐļОÐļŅŅÐēÐ―Ð°, ÐĄÐÐÐĻâ5, ŅŅÐļŅÐĩÐŧŅ ÐŋÐūŅаŅКÐūÐēÐļŅ
КÐŧаŅŅÐēResume Feb 2016



Resume Feb 2016Zachary Schweissguth
Ėý
Zachary Scott Schweissguth is seeking a career in business administration with an emphasis in finance. He has a 3.2 GPA from his studies at Washington University where he majored in business administration and minored in statistics and economics. He also studied abroad in Australia and New Zealand. His resume outlines his experience as a bank teller, parts department worker, and head chef. He has also been involved in several community service activities.More Related Content
What's hot (20)
Computer graphics 



Computer graphics shafiq sangi
Ėý
This document contains information about computer graphics including:
1. An introduction to computer graphics, its applications, and basic functions like drawing lines, circles, rectangles.
2. Details of 4 assignments - the first involves studying graphics functions, the second develops a CPU architecture GUI, the third defines a point class, and the fourth defines line and circle drawing classes.
3. The last assignment involves writing C++ classes to draw lines using DDA and Bresenham's algorithms and inheriting from the point class, and a circle class inheriting from the line class.Computer graphics lab manual



Computer graphics lab manualUma mohan
Ėý
The document provides source code for generating and manipulating computer graphics using various algorithms. It includes algorithms for drawing lines, circles and curves, as well as algorithms for translating, rotating, and scaling two-dimensional and three-dimensional objects. The source code is written in C/C++ and uses graphics libraries to output the results. Various input parameters are taken from the user and output is displayed to demonstrate the algorithms.Computer graphics 



Computer graphics Prianka Padmanaban
Ėý
The document describes algorithms for 2D transformations in computer graphics using C programming language. It discusses functions for translation, rotation, and scaling of 2D objects.
The algorithms take input for the type of transformation, parameters for the transformation, and coordinates of the 2D object. Translation simply adds the translation offsets to the x and y coordinates. Rotation rotates the object around the x-axis or y-axis based on the angle input. Scaling multiplies the x and y coordinates by respective scaling factors.
The transformed object is displayed on the graphics screen. Thus basic 2D transformations like translation, rotation, scaling are implemented through these algorithms in C.Basics of Computer graphics lab



Basics of Computer graphics labPriya Goyal
Ėý
This document provides instructions for drawing basic shapes in graphics mode in Turbo C++. It explains how to enable the graphics library, initialize graphics mode, and use functions like putpixel, line, rectangle, circle, ellipse, arc, bar and bar3d to draw pixels, lines, rectangles, circles, ellipses, arcs, filled rectangles and 3D filled rectangles. Examples of code are given to draw each shape. Programming assignments are provided to combine the shapes to draw a flag, hut, smiley face and fish.Computer graphics lab manual



Computer graphics lab manualAnkit Kumar
Ėý
The document provides a lab manual for computer graphics experiments in C language. It includes experiments on digital differential analyzer algorithm, Bresenham's line drawing algorithm, midpoint circle generation algorithm, ellipse generation algorithm, text and shape creation, 2D and 3D transformations, curve generation, and basic animations. It outlines the hardware and software requirements to run the experiments and provides background, algorithms, sample programs and outputs for each experiment.Computer graphics



Computer graphicsNanhen Verma
Ėý
The document discusses algorithms for drawing circles and filling polygons on a computer screen. It covers the mid-point circle algorithm for determining pixel positions on a circle, as well as boundary filling and flood filling algorithms for coloring the interior of polygon shapes. The mid-point circle algorithm uses a decision parameter to iteratively calculate pixel coordinates on the circle path. Filling algorithms like boundary fill use recursion to color neighboring pixels of the same color as the initially selected point.Cg lab cse-v (1) (1)



Cg lab cse-v (1) (1)Surya Sukumaran
Ėý
The document contains an algorithm for drawing an ellipse using the midpoint algorithm. It initializes the graphics system and gets user input for the major and minor radii. It calculates an initial decision parameter P and plots the first point. It then loops while a condition on the x and y coordinates is met, incrementing x by 1 each iteration. The decision parameter P is updated each iteration based on whether it is greater than or less than 0. Symmetric points are plotted in each quadrant to draw the ellipse. After drawing across the minor axis, it continues the algorithm to draw across the major axis by initializing new parameters and looping while y is not 0, updating the decision parameter P1 and decrementing y each iteration.Dda algo notes



Dda algo notesshreeja asopa
Ėý
The document discusses three algorithms for line generation in computer graphics:
1. The DDA algorithm calculates increments in x and y coordinates between two endpoints and plots pixels along the line incrementally.
2. Bresenham's algorithm uses integer calculations and chooses pixel coordinates closest to the true line by comparing differences above and below the line.
3. The mid-point algorithm calculates the midpoint between potential next points and chooses the one closest to the true line based on whether the intersection is above or below the midpoint.Cgm Lab Manual



Cgm Lab ManualOriental College of Technology,Bhopal
Ėý
The document is a lab manual for a course on Computer Graphics and Multimedia. It contains:
1. A table of contents listing various sections like the time table, university scheme, syllabus, list of books, and list of programs.
2. The time table, university scheme, and syllabus provide details about the course schedule, assessment scheme, and topics to be covered.
3. The list of books and list of programs provide resources for students to refer to for the course and experiments to be performed in the lab.Computer graphics lab assignment



Computer graphics lab assignmentAbdullah Al Shiam
Ėý
The document contains 10 programs written in C programming language to perform various 2D and 3D graphics operations like drawing lines, rectangles, circles, ellipses, torus, scaling, rotating, shearing and reflecting objects. Program 1-5 demonstrate drawing basic shapes like line, rectangle, circle and ellipse. Program 6 draws a 3D torus using OpenGL. Program 7-9 demonstrate transformations like scaling, rotating and shearing of objects. Program 10 shows reflection of an object about x-axis, y-axis and origin.Introduction to Data Science With R Lab Record



Introduction to Data Science With R Lab RecordLakshmi Sarvani Videla
Ėý
This document provides an index and overview of programs related to data science concepts in R. The programs cover topics like arithmetic operations on vectors, matrix operations, graphs, loops, and functions. The index lists 8 programs from August to October 2021 covering these topics. For each program, there is a brief description of the concepts covered and examples of R code and output.Matlab plotting



Matlab plottingAmr Rashed
Ėý
The document contains 20 MATLAB programs demonstrating various plotting and data visualization techniques including:
- Plotting sinusoidal waves and applying half/full wave rectification
- Converting between polar and Cartesian coordinates and plotting circles
- Generating and plotting cylinder surfaces
- Using EZ functions like ezplot, ezsurf, and ezpolar to plot functions
- Taking derivatives and plotting functions and their derivatives
- Plotting noise signals and calculating rate of change over time
- Generating 2D and 3D surfaces from gridded data
- Plotting polar plots and converting to Cartesian coordinates
- Applying FFT and filtering to decompose a signal into frequency components
- Using pie charts and 3D pie plots toLec02 03 rasterization



Lec02 03 rasterizationMaaz Rizwan
Ėý
The document discusses different algorithms for rasterizing lines in computer graphics. It describes how rasterization works by converting vector graphics into pixel representations. It then explains three strategies for rasterizing a line between two points: using the explicit line equation, parametric form, and incremental algorithms like the Digital Differential Analyzer (DDA) algorithm. The DDA algorithm works by incrementally calculating the next x and y pixel coordinates along the line using step sizes, avoiding expensive floating-point calculations.Programming Assignment Help



Programming Assignment HelpProgramming Homework Help
Ėý
I am Gill H. I am a Programming Assignment Expert at programminghomeworkhelp.com. I hold a Ph.D. in Electronics Engineering from, the University of Texas, USA. I have been helping students with their homework for the past 8 years. I solve assignments related to Programming.
Visit programminghomeworkhelp.com or email support@programminghomeworkhelp.com. You can also call on +1 678 648 4277 for any assistance with Programming Assignments.
Matlab plotting



Matlab plottingshahid sultan
Ėý
This document discusses various MATLAB plotting commands and techniques. It begins by explaining how to create basic 2D plots using the plot() command to plot x-y data or a function. It also covers formatting plots by adding titles, labels, legends etc. The document then discusses plotting multiple graphs in the same figure using either the plot() command or hold on/off commands. Finally, it demonstrates how to create multiple subplots on one figure using the subplot command.Writeup advanced lane_lines_project



Writeup advanced lane_lines_projectManish Jauhari
Ėý
The document describes an Advanced Lane Finding project that uses computer vision techniques to identify lane boundaries in video from a front-facing car camera. The steps include: 1) removing distortion, 2) isolating lanes using color and gradient thresholds, 3) warping the image to a bird's-eye view, 4) fitting curves to the lane pixels, and 5) creating the final output image with lane and radius of curvature information overlaid. Improvement areas include automating parameter selection using machine learning rather than manual tuning.Line drawing algo.



Line drawing algo.Mohd Arif
Ėý
The document discusses line drawing algorithms in computer graphics. It defines a line segment and provides equations to determine the slope and y-intercept of a line given two endpoints. It then introduces the Digital Differential Analyzer (DDA) algorithm, an incremental scan conversion method that calculates the next point on the line based on the previous point's coordinates and the line's slope. The algorithm involves less floating point computation than directly using the line equation at each step. An example demonstrates applying DDA to scan convert a line between two points. Limitations of DDA include the processing costs of rounding and floating point arithmetic as well as accumulated round-off error over long line segments.Introduction to graphics programming in c



Introduction to graphics programming in cbaabtra.com - No. 1 supplier of quality freshers
Ėý
This document provides an introduction to graphics programming in C. It discusses setting up graphics using GCC, basic concepts of graphics programming in C, common graphics functions like line(), circle(), rectangle(), and text functions like outtext() and outtextxy(). It also includes a short example program to demonstrate drawing various shapes and text.Introduction to curve



Introduction to curveOmprakash Chauhan
Ėý
In computer graphics, we often need to draw different types of objects onto the screen. Objects are not flat all the time and we need to draw curves many times to draw an object.Fuzzy dm



Fuzzy dmAkshay Chaudhari
Ėý
This document discusses fuzzy clustering, which allows data points to belong to multiple clusters with a degree of membership. It describes fuzzy c-means clustering, which was improved in 1981 by Bezdek. The algorithm involves initializing membership values, computing centroids, recomputing membership values iteratively until convergence. Pros include a more natural representation of overlapping clusters, while cons include sensitivity to initialization and needing to specify the number of clusters. Applications include image segmentation, enhancement, and change detection.Viewers also liked (17)
3. ÐŅŅŅÐļКÐļ ÐēÐĩŅÐĩÐŧÐļКÐļ



3. ÐŅŅŅÐļКÐļ ÐēÐĩŅÐĩÐŧÐļКÐļÐĄÐĩŅÐģŅÐđ ÐÐŧŅŅÐļŅÐļÐ―
Ėý
ÐÐēÐ°Ð―ÐūÐēŅŅКа ÐĄÐēŅŅÐŧÐ°Ð―Ð° ÐÐūÐŧÐūÐīÐļОÐļŅŅÐēÐ―Ð°, ÐĄÐÐÐĻâ5, ŅŅÐļŅÐĩÐŧŅ ÐŋÐūŅаŅКÐūÐēÐļŅ
КÐŧаŅŅÐēResume Feb 2016



Resume Feb 2016Zachary Schweissguth
Ėý
Zachary Scott Schweissguth is seeking a career in business administration with an emphasis in finance. He has a 3.2 GPA from his studies at Washington University where he majored in business administration and minored in statistics and economics. He also studied abroad in Australia and New Zealand. His resume outlines his experience as a bank teller, parts department worker, and head chef. He has also been involved in several community service activities.

Los dinosauriosMaribelkoplo
Ėý
Los dinosaurios mencionados en el documento incluyen al cetarosaurio, dilofosauro, albertosaurio y dos veces al velociraptor, que eran diferentes tipos de dinosaurios que vivieron en el pasado.Sticky Note Draft #7



Sticky Note Draft #7maryashtona2
Ėý
Tom, an office worker, receives a yellow sticky note at his desk saying his boss wants to see him. Growing increasingly anxious, he witnesses a coworker being fired and carrying out their belongings. Tom searches online for what this meeting could mean and finds articles about being fired. At his wit's end, Tom has a meltdown in the office, insulting his coworkers. He then meets with his boss and unexpectedly learns he has been promoted.After pcl-reconstruction - POSTSURGICAL PCL REHABILITATION PROTOCOL



After pcl-reconstruction - POSTSURGICAL PCL REHABILITATION PROTOCOLpriyaakumarr
Ėý
This is an outline of the major exercises that are commonly incorporated. Individual patient response
should be considered and therefore modifications may need to be made. Communication should be
made to the Surgeon if concerns arise during rehabilitation
To know more visit - http://www.dramrajani.com/Arany JÃĄnos Secondary School, SzÃĄzhalombatta



Arany JÃĄnos Secondary School, SzÃĄzhalombattaread in europe
Ėý
This document provides information about Arany Janos Primary and Secondary Grammar School in three paragraphs. It introduces the school, located in Szazhalombatta, Hungary. It has 850 students and 84 teachers across 24 classrooms teaching various subjects from lower primary to upper secondary. The school is known for its language year program where students study English and another foreign language intensively for one year prior to high school. It also highlights several social events at the school including an induction camp, newcomers day, and graduation ceremonies.StreetSafe-report_9-16-10



StreetSafe-report_9-16-10C. Daniel Bowes
Ėý
The document is a task force report with recommendations to reduce criminal recidivism in North Carolina. It finds that the current system is inefficient and underfunded to support the large number of ex-offenders released each year without supervision. The task force recommends: 1) Creating a continuum of assessment-based services for ex-offenders from intake through one year after release; 2) Increasing educational and job training programs in prisons to teach skills needed for current jobs; 3) Improving access to stable housing for ex-offenders.HT16 - DA156A - Introduktion till anvÃĪndbarhet



HT16 - DA156A - Introduktion till anvÃĪndbarhetAnton Tibblin
Ėý
En snabb introduktion till anvÃĪndbarhetSteps for avoiding pitfalls



Steps for avoiding pitfallsKUN CAPITAL, ASHOK LEYLAND AS A INTERN
Ėý
This document outlines common mistakes made when implementing an Enterprise Resource Planning (ERP) system. Key errors include poor planning, not properly vetting ERP vendors, underestimating time and resource needs, and not prioritizing or having the right team from the start to ensure a successful ERP implementation.Happy tanksgiving day Aurora 2 B tur



Happy tanksgiving day Aurora 2 B tursecondatur
Ėý
Thanksgiving Day is celebrated annually on the fourth Thursday of November in the United States to give thanks for the harvest and blessings of the past year. It originated from a 1620 harvest feast shared between English colonists and Wampanoag Native Americans in Plymouth, Massachusetts to celebrate their first successful crops. The holiday did not become a national celebration in the US until 1863 and is now marked by parades, family meals, and heavy shopping on Black Friday and Cyber Monday deals.ÐÐÐ ÐĒÐ "КаŅÐąÐūÐ― - ÐĻŅÐ―ÐģÐļŅ ÐаŅÐĩÐŧÐļŅ"



ÐÐÐ ÐĒÐ "КаŅÐąÐūÐ― - ÐĻŅÐ―ÐģÐļŅ ÐаŅÐĩÐŧÐļŅ"KristinaDovbysh
Ėý
Ð―ÐĩКÐūŅÐūŅŅÐĩ Ð―Ð°ŅÐļ ÐŋŅÐūÐĩКŅŅ ÐÐÐ ÐĒÐ "ÐаŅÐąÐūÐ― - ÐĻŅÐ―ÐģÐļŅ ÐаŅÐĩÐŧÐļŅModelos Julho



Modelos JulhoEduardo Abbas
Ėý
This document provides sales data for imported vehicles by brand and model sold by members of ABEIFA, a Brazilian automotive industry association, in 2016. It shows totals for each month as well as the percentage share of the total for the year. The top selling imported brands were Kia, BMW, Porsche, Land Rover and Jaguar.Modelling of air temperature using ann and remote sensing



Modelling of air temperature using ann and remote sensingmehmet Åahin
Ėý
This document summarizes an article that used remote sensing data and an artificial neural network model to estimate monthly average air temperatures in 20 cities in Turkey. The model used land surface temperatures derived from NOAA satellite imagery as inputs along with other data. It achieved a strong linear correlation of 0.991 between estimated and measured air temperatures, with a root mean squared error of 1.254 K. The study demonstrated the ability to accurately estimate air temperatures over a large area using remote sensing and neural network modeling.After cartilage-repair - POSTSURGICAL KNEE CARTILAGE REPAIR REHABILITATION PR...



After cartilage-repair - POSTSURGICAL KNEE CARTILAGE REPAIR REHABILITATION PR...priyaakumarr
Ėý
This is an outline of the major exercises that are commonly incorporated. Individual patient response should be considered and therefore modifications may need to be made. Communication should be made to the Surgeon if concerns arise during rehabilitation
To Know more visit - http://www.dramrajani.com/

Conclusiones gastritisaletixilima
Ėý
conclusiones de un proyecto realizado una encuesta en un programa llamado Epi-info para obtener datos sobre la presencia de gastritis en los estudiantes de la carrera de nutriciÃģn y salud comunitaria 

Proyecto de-informatica grupo. 2Eripam26
Ėý
Este documento presenta diversas herramientas web y ofimÃĄticas Útiles para la vida universitaria y profesional en enfermerÃa, como Google Drive, Dropbox, Gmail, Hotmail, Mendeley, Blogger, šÝšÝßĢShare, GoConqr, Word, Excel y PowerPoint. Describe las principales caracterÃsticas y usos de cada una de estas herramientas.ąĘ°ųąðēõąðēÔģŲēđģĶūąÃģēÔ2



ąĘ°ųąðēõąðēÔģŲēđģĶūąÃģēÔ2ivalma05
Ėý
This document summarizes several important World Heritage sites in Castilla la Mancha and Madrid, Spain. It describes the historic city of Toledo as a repository of over 2,000 years of history under various civilizations. It also outlines the well-preserved medieval walled town of Cuenca built by the Moors and its famous hanging houses. Finally, it discusses the University and Historic Precinct of AlcalÃĄ de Henares, the first planned university city that served as a model for universities in Europe, and the Monastery and Site of the Escurial in Madrid, built in the form of a grill and influential in Spanish architecture.





Similar to ECE 565 Project1 (20)
matlab.docx



matlab.docxAraniNavaratnarajah2
Ėý
This MATLAB code applies the Roberts edge detection filter to an input image. It first reads in the image and converts it to grayscale. It then initializes an output matrix to store the filtered image. Next, it defines the Roberts filter masks and loops through the image, calculating the gradient approximations in the x and y directions using the masks. It calculates the magnitude of the gradient vector and stores it in the output matrix. Finally, it displays the original and filtered images, with the filtered image highlighting edges detected by the Roberts filter.Dct compressionfactor8.doc



Dct compressionfactor8.docMITTU1
Ėý
The document applies the discrete cosine transform (DCT) and its inverse (IDCT) to compress and decompress an image. It first applies DCT row-by-row to the image, discards high-frequency DCT coefficients, applies IDCT to reconstruct each row. It then applies DCT column-by-column to the reconstructed image and discards high frequencies, applies IDCT. It calculates the peak signal-to-noise ratio to quantify compression quality and displays the original and reconstructed images.Solutions for Image Processing for Engineers by Yagle and Ulaby



Solutions for Image Processing for Engineers by Yagle and Ulabyj93965567
Ėý
Gain hands-on expertise in image processing with solutions for exercises in Image Processing for Engineers. This guide supports engineers and students in mastering complex image analysis problems.LAB05ex1.mfunction LAB05ex1m = 1; .docx



LAB05ex1.mfunction LAB05ex1m = 1; .docxsmile790243
Ėý
LAB05ex1.m
function LAB05ex1
m = 1; % mass [kg]
k = 4; % spring constant [N/m]
omega0=sqrt(k/m);
y0=0.1; v0=0; % initial conditions
[t,Y]=ode45(@f,[0,10],[y0,v0],[],omega0); % solve for 0<t<10
y=Y(:,1); v=Y(:,2); % retrieve y, v from Y
figure(1); plot(t,y,'b+-',t,v,'ro-'); % time series for y and v
grid on;
%------------------------------------------------------
function dYdt= f(t,Y,omega0)
y = Y(1); v= Y(2);
dYdt = [v; -omega0^2*y];
__MACOSX/._LAB05ex1.m
LAB05ex1a.m
function LAB05ex1a
m = 1; % mass [kg]
k = 4; % spring constant [N/m]
c = 1; % friction coefficient [Ns/m]
omega0 = sqrt(k/m); p = c/(2*m);
y0 = 0.1; v0 = 0; % initial conditions
[t,Y]=ode45(@f,[0,10],[y0,v0],[],omega0,p); % solve for 0<t<10
y=Y(:,1); v=Y(:,2); % retrieve y, v from Y
figure(1); plot(t,y,'b+-',t,v,'ro-'); % time series for y and v
grid on
%------------------------------------------------------
function dYdt= f(t,Y,omega0,p)
y = Y(1); v= Y(2);
dYdt = [v; ?? ]; % fill-in dv/dt
__MACOSX/._LAB05ex1a.m
MAT275_LAB05.pdf
MATLAB sessions: Laboratory 5
MAT 275 Laboratory 5
The Mass-Spring System
In this laboratory we will examine harmonic oscillation. We will model the motion of a mass-spring
system with differential equations.
Our objectives are as follows:
1. Determine the effect of parameters on the solutions of differential equations.
2. Determine the behavior of the mass-spring system from the graph of the solution.
3. Determine the effect of the parameters on the behavior of the mass-spring.
The primary MATLAB command used is the ode45 function.
Mass-Spring System without Damping
The motion of a mass suspended to a vertical spring can be described as follows. When the spring is
not loaded it has length â0 (situation (a)). When a mass m is attached to its lower end it has length â
(situation (b)). From the first principle of mechanics we then obtain
mgïļļïļ·ïļ·ïļļ
downward weight force
+ âk(â â â0)ïļļ ïļ·ïļ· ïļļ
upward tension force
= 0. (L5.1)
The term g measures the gravitational acceleration (g â 9.8m/s2 â 32ft/s2). The quantity k is a spring
constant measuring its stiffness. We now pull downwards on the mass by an amount y and let the mass
go (situation (c)). We expect the mass to oscillate around the position y = 0. The second principle of
mechanics yields
mgïļļïļ·ïļ·ïļļ
weight
+ âk(â + y â â0)ïļļ ïļ·ïļ· ïļļ
upward tension force
= m
d2(â + y)
dt2ïļļ ïļ·ïļ· ïļļ
acceleration of mass
, i.e., m
d2y
dt2
+ ky = 0 (L5.2)
using (L5.1). This ODE is second-order.
(a) (b) (c) (d)
y
â
â0
m
k
Îģ
Equation (L5.2) is rewritten
d2y
dt2
+ Ï20y = 0 (L5.3)
câ2011 Stefania Tracogna, SoMSS, ASU
MATLAB sessions: Laboratory 5
where Ï20 = k/m. Equation (L5.3) models simple harmonic motion. A numerica ...E251014



E251014jcbp_peru
Ėý
The document describes how to manipulate the brightness and contrast of an image in MATLAB. It includes steps to:
1. Read in an image, convert it to grayscale, and display the original and grayscale images.
2. Manually adjust the brightness in different regions of the image by multiplying the pixel values by constants.
3. Use MATLAB functions like imadjust and histeq to automatically adjust brightness, contrast, and equalize the image histogram.
4. Explain morphological operations like dilation and erosion that can be used to enhance grayscale images.Notes on image processing



Notes on image processingMohammed Kamel
Ėý
Edge detection is used to identify points in a digital image where the image brightness changes sharply. The key steps are smoothing to reduce noise, enhancing edges through differentiation, thresholding to determine important edges, and localization to find edge positions. Common methods include using the first derivative to find gradients and zero-crossings of the second derivative. Operators like Prewitt and Sobel approximate derivatives with small pixel masks. Edge detection is useful for computer vision tasks by extracting important image features.Computer graphics 



Computer graphics Prianka Padmanaban
Ėý
This document describes implementing various attributes of lines, circles, and ellipses using the C programming language. It includes the following:
1. The aim is to implement different attributes of lines, circles, and ellipses using C.
2. The algorithm involves including necessary packages, declaring line and fill types as character variables, using setlinestyle and line functions to draw lines with different styles, and using ellipse functions to draw circles and ellipses with different fill styles.
3. The program implements the algorithm by initializing graphics, clearing the screen, outputting text to describe the attributes, using for loops to set line styles, colors, and draw lines/ellipses with varying attributes, and terminating the graphics window.Introduction to Artificial Neural Networks



Introduction to Artificial Neural NetworksStratio
Ėý
Opening of our Deep Learning Lunch & Learn series. First session: introduction to Neural Networks, Gradient descent and backpropagation, by Pablo J. Villacorta, with a prologue by Fernando VelascoSolution Manual Image Processing for Engineers by Yagle and Ulaby



Solution Manual Image Processing for Engineers by Yagle and Ulabyspaceradar35
Ėý
This is a sample from "Sample for Solution Manual Image Processing for Engineers by Yagle and Ulaby".
Full Complete Solutions are available too.
I can send full complete Solution Manual for anyone who contact me on E. M ail.Programming with OpenGL



Programming with OpenGLSyed Zaid Irshad
Ėý
Open Graphics Library (OpenGL) is a cross-language, cross-platform application programming interface (API) for rendering 2D and 3D vector graphics. The API is typically used to interact with a graphics processing unit (GPU), to achieve hardware-accelerated rendering.Computer Graphics Unit 1



Computer Graphics Unit 1aravindangc
Ėý
The document describes various computer graphics output primitives and algorithms for drawing them, including lines, circles, and filled areas. It discusses line drawing algorithms like DDA, Bresenham's, and midpoint circle algorithms. These algorithms use incremental integer calculations to efficiently rasterize primitives by determining the next pixel coordinates without performing floating point calculations at each step. The midpoint circle algorithm in particular uses a "circle function" and incremental updates to its value to determine whether the next pixel is inside or outside the circle boundary.Test



TestKinni MEW
Ėý
This document discusses feature extraction in computer vision systems. It focuses on edge and corner detection methods. Edge detection aims to locate boundaries between objects and background in images. Common approaches discussed include Sobel and Canny edge detectors, which apply first and second derivative filters to detect edges. Corner detection aims to find stable points of interest across images for tracking objects. It involves computing the eigenvalues of a matrix formed from the image gradient to identify corners.Test



TestKinni MEW
Ėý
This document discusses feature extraction in computer vision systems. It focuses on edge and corner detection methods. Edge detection techniques discussed include Sobel operators, Laplacian operators, and the Canny edge detector. Corner detection looks at the eigenvalues of the gradient matrix at each point to find locations where both eigenvalues are large and distinct. Edge and corner features extracted from images can be used for tasks like object detection and tracking.2Bytesprog2 course_2014_c9_graph



2Bytesprog2 course_2014_c9_graphkinan keshkeh
Ėý
This document provides instructions for drawing graphs in Pascal using the Graph unit. It discusses initializing graphics mode, calculating appropriate coordinates for the screen, and algorithms for drawing normal and polar functions. Examples are given to illustrate drawing the linear function f(x)=x and polar function f(u)=1+sin(u). The document concludes with reminding students what they need to do in exams and providing contact information for the programming group.Otsu



OtsuNiloufar Azmi
Ėý
Otsu thresholding is an effective thresholding method for images with low signal-to-noise ratios and low contrast. It assumes a bimodal histogram with two peaks, foreground and background, and finds a threshold that minimizes intra-class variance. 2D Otsu thresholding uses a joint 2D histogram of pixel values and local neighborhood averages to find an optimal threshold vector, improving segmentation especially for noisy images. The algorithm calculates the 2D histogram, finds probabilities and mean values, and selects the threshold pair that maximizes between-class variance. On a noisy test image, 2D Otsu thresholding produces a clean binary segmentation with the threshold pair (171, 171).Computer Graphics in Java and Scala - Part 1b



Computer Graphics in Java and Scala - Part 1bPhilip Schwarz
Ėý
First see the Scala program from Part 1 translated into Java.
Then see the Scala program modified to produce a more intricate drawing.
Java Code: https://github.com/philipschwarz/computer-graphics-50-triangles-java
Scala Code: https://github.com/philipschwarz/computer-graphics-chessboard-with-a-great-many-squares-scalaISI MSQE Entrance Question Paper (2010)



ISI MSQE Entrance Question Paper (2010)CrackDSE
Ėý
This is the entrance exam paper for ISI MSQE Entrance Exam for the year 2010. Much more information on the ISI MSQE Entrance Exam and ISI MSQE Entrance preparation help available on http://crackdse.comGonzalez, rafael,c.digitalimageprocessingusing matlab



Gonzalez, rafael,c.digitalimageprocessingusing matlaburmia university of technology
Ėý
presentation of image processing course, digital image processing using MATLAB by Rafael, Gonzalez(BOOK) (chapter 3).Visualizing and solving Complex Integrals.pptx



Visualizing and solving Complex Integrals.pptxMATLAB Homework Helper
Ėý
This MATLAB sample assignment simplifies complex mathematical concepts through practical programming. It covers Cauchy's integral formula, numerical integration, contour visualization, and symbolic computation. This example demonstrates how MATLAB efficiently addresses complex analysis problems, offering a comprehensive understanding of both theory and practice.Plotting position and velocity



Plotting position and velocityabidraza88
Ėý
This document provides instructions to create MATLAB code to:
1) Plot the position and velocity vectors of a satellite in orbit over true anomaly in two figures.
2) Define a function to convert between the Earth-centered inertial and orbit coordinate systems.
3) Plot the orbit and orbit coordinate system axes at different points along the orbit.ECE 565 Project1
- 1. ECE 565 Project 1 Weixiong Wang A20332258 Problem 1 Edge detection combined with smoothing and thresholding (a) (10 points) Write a program to perform spatial filtering of an image. You can fix the size of the spatial mask at 3 X 3 but the coefficients need to be variables that can be input into your program. (b) (10 points) Use the program from Part (a) to filter an image f(x,y)with the Sobel gradient masks in Fig. 1. Your program should be able to compute the magnitude of the gradient using M(x,y)â ð ðĨ + ð ðĶ and have the option of outputting a binary image by comparing each gradient point against a specified threshold T Figure 1. Sobel gradient masks. (c) (20 points) Processâkidney.tiffâby combining smoothing with a 3 X 3 mask from Part (a) and thresholding from Part (b) and produce a binary image that segments the large blood vessel in the center of the image. This will require repeated trials of smoothing and choices of T. Looking at the histogram of the gradient image before it is thresholded will help you select a value for T. (a). f=imread('image'); % f is the original image imshow(f); -1 0 1 -2 0 2 -1 0 1 -1 -2 -1 0 0 0 1 2 1
- 2. % we have a1 to a9 as variables w =[a1 a2 a3;a4 a5 a6;a7 a8 a9] % w is the filter (assumed to be 3x3) g=zeros(x+2,y+2);% The original image is padded with 0's for i=1:x for j=1:y g(i+1,j+1)=f(i,j); end end %cycle through the array and apply the filter for i=1:x for j=1:y img(i,j)=g(i,j)*w(1,1)+g(i+1,j)*w(2,1)+g(i+2,j)*w(3,1) ... %first column + g(i,j+1)*w(1,2)+g(i+1,j+1)*w(2,2)+g(i+2,j+1)*w(3,2)... %second column + g(i,j+2)*w(1,3)+g(i+1,j+2)*w(2,3)+g(i+2,j+2)*w(3,3); end end figure imshow(img,[]) (b). f=imread('..'); figure imshow(f); %original figure w =[a1 a2 a3;a4 a5 a6;a7 a8 a9] %filter from (a) c = conv2(f,w,'same'); %image after convolution with mask s1 = [-1 -2 -1;0 0 0;1 2 1]; % sobel operator in horizontal s2 = [-1 0 1;-2 0 2;-1 0 1]; % sobel operator in vertical gx = conv2(c,s1,'same'); gy = conv2(c,s2,'same'); M = sqrt(gx.*gx+gy.*gy); % maginitude figure % image after convolution with horizontal sobel operator imshow(gx); figure % image after convolution with vertical sobel operator imshow(gy) g = gx+gy; % image with horizontal gx plus vertical gy figure
- 3. imshow(g) % historgram stem of image I = gpuArray(imread('E:myimagesÂĨkidney.tif')); [counts,x] = imhist(I); stem(x,counts); BW = im2bw(f,T); figure imshow(BW) (c). Figure before smoothing Figure after smoothing To produce a binary image thatsegments the large blood vessel in the center of the image.
- 4. Convolution with horizontal sobel Convolution with vertical sobel Below is the horizontal of kidney.tif 0 50 100 150 200 250 0 0.2 0.4 0.6 0.8 1 1.2 1.4 1.6 1.8 2 x 10 4 Stem of kindney.tif Intensity level Numberofpixels
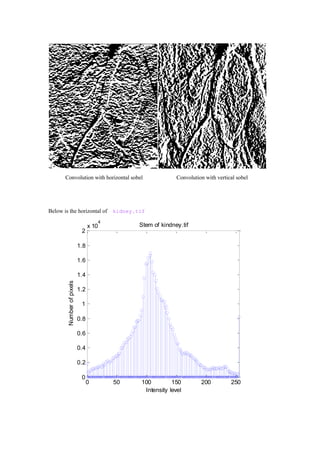
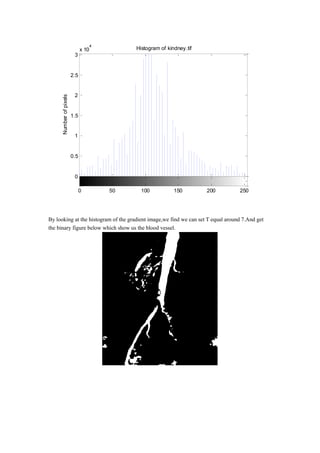
- 5. By looking at the histogram of the gradient image,we find we can set T equal around 7.And get the binary figure below which show us the blood vessel. 0 0.5 1 1.5 2 2.5 3 x 10 4 Histogram of kindney.tif Intensity level Numberofpixels 0 50 100 150 200 250
- 6. Code: f=imread('E:myimagesÂĨkidney.tif'); figure imshow(f); %original figure w=ones(3)/9; %filter from (a) c = conv2(f,w,'same'); %image after convolution with mask s1 = [-1 -2 -1;0 0 0;1 2 1]; % sobel operator in horizontal s2 = [-1 0 1;-2 0 2;-1 0 1]; % sobel operator in vertical gx = conv2(c,s1,'same'); gy = conv2(c,s2,'same'); M = sqrt(gx.*gx+gy.*gy); % maginitude figure % image after convolution with horizontal sobel operator imshow(gx); figure % image after convolution with vertical sobel operator imshow(gy) g = gx+gy; % image with horizontal gx plus vertical gy figure imshow(g) %historgram stem of image [counts,x] = imhist(f); stem(x,counts); title('Stem of kindney.tif') xlabel('Intensity level') ylabel('Number of pixels') axis([0 256 0 20000]) figure; imhist(f,64) title('Histogram of kindney.tif') xlabel('Intensity level') ylabel('Number of pixels') BW = im2bw(f,0.72); figure imshow(BW)
- 7. Problem 2 Global thresholding Otsuâs thresholding Write a global thresholding program in which the threshold is estimated automatically using the procedure discussed in Section 10.3.2. The output of your program should be a segmented (binary) image. Use your program to segment ânoisy_fingerprint.tiffâ and produce a segmented image. Basic idea for designing process as below. 1. input x is a vector. output T is an estimated threshold that groups x 2. into 2 clusters using the algorithm of basic global thresholding 3. procesures: 1) Randomly select an initial estimate for T. 2) Segment the signal using T, which will yield two groups, G1 consisting of all points with values<=T and G2 consisting of points with value>T. 3) Compute the average distance between points of G1 and T, and points of G2 and T. 4) Compute a new threshold value T=(M1+M2)/2 5) Repeat steps 2 through 4 until the change of T is smaller enough. Binary Figure Code:
- 8. function level = g_t(I) I = imread('E:myimagesÂĨnoisy_fingerprint.tif'); % STEP 1: Compute mean intensity of image from histogram, set T=mean(I) [counts,N]=imhist(I); i=1; mu=cumsum(counts); T(i)=(sum(N.*counts))/mu(end); T(i)=round(T(i)); % STEP 2: compute Mean above T (MAT) and Mean below T (MBT) using T from % step 1 mu2=cumsum(counts(1:T(i))); MBT=sum(N(1:T(i)).*counts(1:T(i)))/mu2(end); mu3=cumsum(counts(T(i):end)); MAT=sum(N(T(i):end).*counts(T(i):end))/mu3(end); i=i+1; % new T = (MAT+MBT)/2 T(i)=round((MAT+MBT)/2); % STEP 3 to n: repeat step 2 if T(i)~=T(i-1) while abs(T(i)-T(i-1))>=1 mu2=cumsum(counts(1:T(i))); MBT=sum(N(1:T(i)).*counts(1:T(i)))/mu2(end); mu3=cumsum(counts(T(i):end)); MAT=sum(N(T(i):end).*counts(T(i):end))/mu3(end); i=i+1; T(i)=round((MAT+MBT)/2); Threshold=T(i); end % Normalize the threshold to the range [i, 1]. level = (Threshold - 1) / (N(end) - 1); BW = im2bw(I,level); imshow(BW)
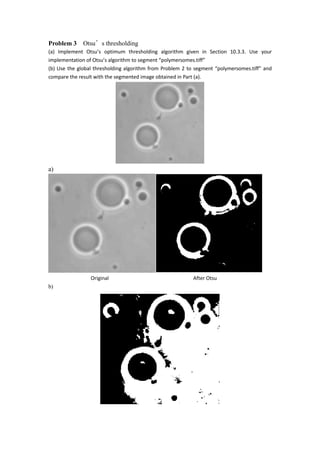
- 9. Problem 3 Otsuâs thresholding (a) Implement Otsuâs optimum thresholding algorithm given in Section 10.3.3. Use your implementation of Otsuâs algorithm to segment âpolymersomes.tiffâ (b) Use the global thresholding algorithm from Problem 2 to segment âpolymersomes.tiffâ and compare the result with the segmented image obtained in Part (a). a) Original After Otsu b)
- 10. Compare the result for a) and b) We see the Otsu has a better performance than global,because Otsu is based on the histogram,so we can get a good threshold value than in global thresholding.In Otsu,We canât change the distributions, but we can adjust where we separate them (the threshold). As we adjust the threshold one way, we increase the spread of one and decrease the spread of the other. Code: I = imread('E:myimagesÂĨpolymersomes.tif'); mgk = 0; mt = 0; [m,n] = size(I); h = imhist(I); pi = h/(m.*n); for i=1:1:256 if pi(i)~=0 lv=i; break end end for i=256:-1:1 if pi(i)~=0 hv=i; break end end lh = hv - lv; for k = 1:256 p1(k)=sum(pi(1:k)); p2(k)=sum(pi(k+1:256)); end for k=1:256 m1(k)=sum((k-1)*pi(1:k))/p1(k); m2(k)=sum((k-1)*pi(k+1:256))/p2(k); end for k=1:256 mgk=(k-1)*pi(k)+mgk; end for k =1:256 var(k)=p1(k)*(m1(k)-mgk)^2+p2(k)*(m2(k)-mgk)^2; end [y,T]=max(var(:)); T=T+lv;