Mlpa (Multipleks PCR)
- 2. MLPA tekniÄi âĒ MLPA tek bir nÞklotid deÄiÅikliÄi olan dizilerin dahi ayrÄąmÄąnÄą saÄlayacak Åekilde, 50 kadar farklÄą genomik DNA veya RNA dizisindeki normal olmayan kopya sayÄąsÄąnÄąn tespitini saÄlayan bir multipleks PCR yÃķntemidir. âĒ Ä°lk kez 2002 yÄąlÄąnda (Schouten JP et al. Nucleic Acids Res). âĒ BugÞne kadar MLPA tekniÄi kullanÄąlarak 750âden fazla makale yayÄąnlandÄą. âĒ Her yÄąl 1,000,000â dan fazla çalÄąÅma MLPA ile yapÄąlÄąyor. âĒ DÞnya genelinde 80 Þlkede, 800âden fazla labâda rutin olarak kullanÄąlmakta.
- 3. NOT: MLPA çalÄąÅmasÄą için sadece bir termalcycler ve kapiler elektroforez cihazÄąna ihtiyaç duyulur. AynÄą anda 96 numuneyi 24 saat içinde sonuç alacak Åekilde iÅleme tabi tutmak mÞmkÞndÞr.
- 4. MLPAânÄąn getirdiÄi avantajlar MLPA, kopya sayÄąsÄą belirlenmesi konusunda diÄer tekniklere karÅÄąn daha hassastÄąr. (Dizi analizi (sekans) ve DHPLC gibi yÃķntemler, genellikle kopya sayÄąsÄą deÄiÅikliÄi tespitinde baÅarÄąsÄąz kalmaktadÄąr) Ä°yi karakterize edilmiÅ delesyon ve amplifikasyonlar PCR ile belirlenebilse de, çoÄunlukla delesyonlarÄąn tam kÄąrÄąlma noktalarÄą bilinmemektedir. FISH ile tespiti mÞmkÞn olmayan sÄąk tek gen deÄiÅiklerinin (çok kÄąsa diziler 50-70 nt) belirlenebilmesi saÄlamasÄą bakÄąmÄąndan avantaj sunmaktadÄąr. âArray CGHâ ile karÅÄąlaÅtÄąrdÄąÄÄąmÄązda, MLPA daha az maliyetli ve daha az karmaÅÄąk bir yÃķntemdir. MLPA tÞm genomu hedefleyen araÅtÄąrma taramalarÄą için uygun olmamakla birlikte, birçok rutin uygulama içinâarrayâ tabanlÄą tekniklere iyi bir alternatiftir. 45 farklÄą dizilimi hedefleyen MLPA reaksiyonu, sadece 20 ng insan DNAâsÄą gerektirir (~3000 hÞcre) ve sadece bir nÞkleotitteki farklÄąlÄąÄÄą içeren dizilimleri ayÄąrt edebilme potansiyeline sahiptir.
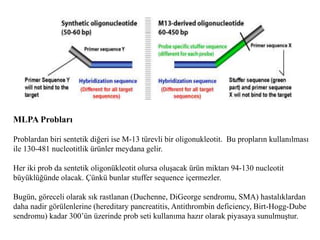
- 5. MLPA ProblarÄą Problardan biri sentetik diÄeri ise M-13 tÞrevli bir oligonukleotit. Bu proplarÄąn kullanÄąlmasÄą ile 130-481 nucleotitlik ÞrÞnler meydana gelir. Her iki prob da sentetik oligonÞkleotit olursa oluÅacak ÞrÞn miktarÄą 94-130 nucleotit bÞyÞklÞÄÞnde olacak. ÃÞnkÞ bunlar stuffer sequence içermezler. BugÞn, gÃķreceli olarak sÄąk rastlanan (Duchenne, DiGeorge sendromu, SMA) hastalÄąklardan daha nadir gÃķrÞlenlerine (hereditary pancreatitis, Antithrombin deficiency, Birt-Hogg-Dube sendromu) kadar 300âÞn Þzerinde prob seti kullanÄąma hazÄąr olarak piyasaya sunulmuÅtur.
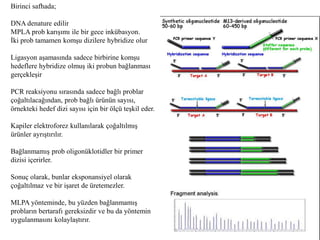
- 6. Birinci safhada; DNA denature edilir MPLA prob karÄąÅÄąmÄą ile bir gece inkÞbasyon. Ä°ki prob tamamen komÅu dizilere hybridize olur Ligasyon aÅamasÄąnda sadece birbirine komÅu hedeflere hybridize olmuÅ iki probun baÄlanmasÄą gerçekleÅir PCR reaksiyonu sÄąrasÄąnda sadece baÄlÄą problar çoÄaltÄąlacaÄÄąndan, prob baÄlÄą ÞrÞnÞn sayÄąsÄą, Ãķrnekteki hedef dizi sayÄąsÄą için bir ÃķlçÞ teÅkil eder. Kapiler elektroforez kullanÄąlarak çoÄaltÄąlmÄąÅ ÃžrÞnler ayrÄąÅtÄąrÄąlÄąr. BaÄlanmamÄąÅ prob oligonÞklotidler bir primer dizisi içerirler. Sonuç olarak, bunlar eksponansiyel olarak çoÄaltÄąlmaz ve bir iÅaret de Þretemezler. MLPA yÃķnteminde, bu yÞzden baÄlanmamÄąÅ problarÄąn bertarafÄą gereksizdir ve bu da yÃķntemin uygulanmasÄąnÄą kolaylaÅtÄąrÄąr.
- 7. MLPA Reaksiyonu MLPA reaksiyonunu beÅ safhaya ayÄąrÄąlabilir ; 1. DNA denaturasyonu (Heat 5 mins at 98°C) 2. MLPA problarÄąnÄąn hibridizasyonu (Incubate 1 minute at 95°C + 16 hours hybridization at 60°C) 3. ligasyon (baÄlanma) reaksiyonu (Add ligase mix and incubate 15 minutes at 54°C; Heat to inactivate the ligase: 5 minutes 98°C) 4. PCR reaksiyonu (Add primers, dNTPs and polymerase; Start PCR) 5. Amplifiye ÞrÞnÞn elektroforez ile ayrÄąÅtÄąrÄąlmasÄą 6. Veri analizi (Determine RELATIVE size of the fluorescent peaks, Compare results to reference samples)
- 8. KullanÄąm alanlarÄą; ïž Mutasyon analizleri ïž Tek nÞkleotit polimorfizim ïž DNA metilasyon analizi ïž mRNA kantitasyonu ïž HÞcre hatlarÄąnÄąn ve dokularÄąn kromozom analizleri ïž Gen kopya sayÄąlarÄąnÄąn analizi ïž Kanser genlerindeki ( BRCA1, BRCA2, hMLH1 ve hMSH2) duplikasyon ve delesyonlarÄąn tespiti ïž AnÃķploidi tespiti ïž Prenetal teÅhisâĶvb.
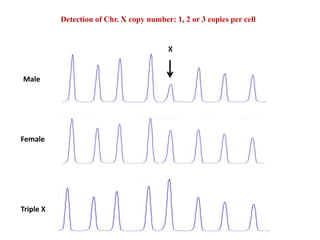
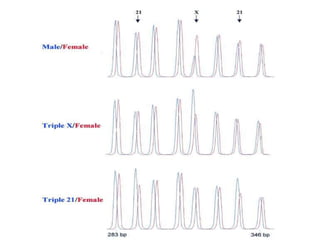
- 9. Detection of Chr. X copy number: 1, 2 or 3 copies per cell X Male Female Triple X
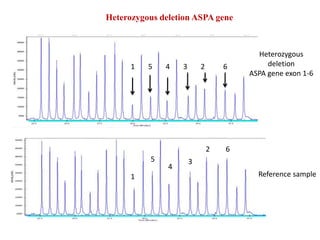
- 12. Heterozygous deletion ASPA gene Heterozygous deletion ASPA gene exon 1-6 Reference sample1 1 5 5 4 4 3 3 2 2 6 6
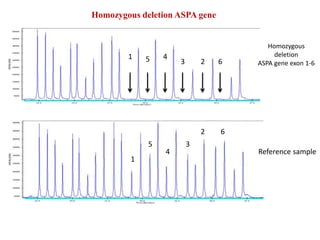
- 13. Homozygous deletion ASPA gene Homozygous deletion ASPA gene exon 1-6 Reference sample 1 5 4 3 2 6 1 5 4 3 2 6
- 14. Major applications of MLPA - mutation detection -
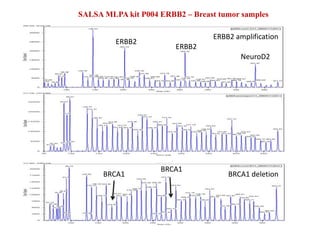
- 15. ERBB2 ERBB2 NeuroD2 SALSA MLPA kit P004 ERBB2 â Breast tumor samples ERBB2 amplification BRCA1 deletionBRCA1 BRCA1
- 16. Methylation detection âĒ Epigenetics â Regulation of gene transcription due to methylation of CpG nucleotides located within the promoters. â Example: Hypermethylation of the MGMT promoter in brain tumors. âĒ Imprinting â Regions of the genome where the methylation patterns are preserved in the same way as they were inherited from the parents. â Examples: The chr.15 Prader-Willy/Angelman region and the chr. 11 Beckwidt-Wiedemann region. Detection of aberrant CpG island methylation.
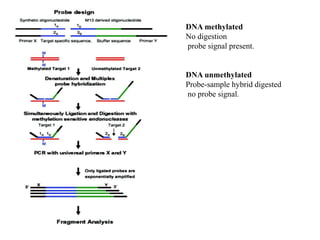
- 17. DNA methylated No digestion probe signal present. DNA unmethylated Probe-sample hybrid digested no probe signal.
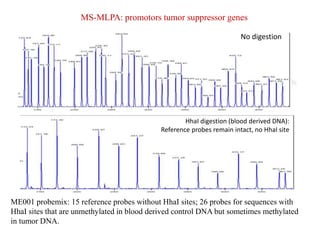
- 18. MS-MLPA: promotors tumor suppressor genes No digestion HhaI digestion (blood derived DNA): Reference probes remain intact, no HhaI site ME001 probemix: 15 reference probes without HhaI sites; 26 probes for sequences with HhaI sites that are unmethylated in blood derived control DNA but sometimes methylated in tumor DNA.
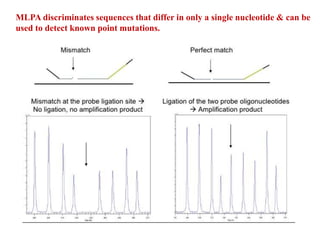
- 19. MLPA discriminates sequences that differ in only a single nucleotide & can be used to detect known point mutations.
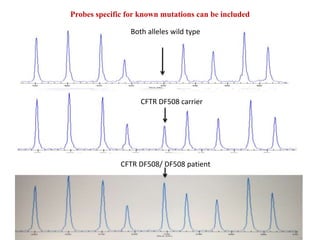
- 20. Probes specific for known mutations can be included CFTR DF508 carrier Both alleles wild type CFTR DF508/ DF508 patient
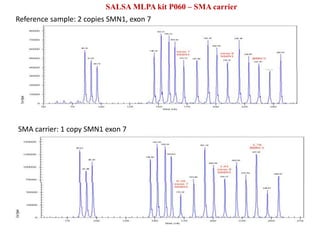
- 21. -Spinal Muscular Dystrophy â âĒ The SMN1 and SMN2 genes are very closely related. âĒ Important difference: one nucleotide change in exon 7. âĒ P021 SMA probemix: â copy number of both SMN1 and SMN2 â used for patient samples. âĒ P060 SMA carrier probemix: â copy number changes of SMN1 â used for carrier screening. âĒ The presence of zero, one, two, three and four copies of SMN1 or SMN2 can be determined. âĒ In 95% of the cases, the cause of SMA can be detected by one MLPA reaction. Major applications of MLPA
- 22. SALSA MLPA kit P060 â SMA carrier Reference sample: 2 copies SMN1, exon 7 SMA carrier: 1 copy SMN1 exon 7