SciScore for Rigor and Reproducibility
- 1. SciScorefor rigor and reproducibility Anita Bandrowski Martijn Roelandse A tool supported by R43OD024432, R44MH119094
- 2. Reproducibility crisis the effect size of a poorly controlled study is about 50% bigger than the effect size of a well controlled study. Is it possible that poorly controlled animal studies are repeated using proper controls in clinical trials and fail because the effects were never significant to begin with?
- 3. Entrez RRID
- 5. Sowhere arewe now? NIF, INCF, members of the NIH, and about 25 major journal Editors in Chief, began to talk about research resource reproducibility ŌĆó 2012: 1st meeting at the Commander's Palace @ Society for Neuroscience 2013: 2nd meeting at NIH ŌĆó 2014: Pilot project started; 25 journals would ask authors to provide RRIDs for 3 months, 2 journals started on time we are currently in 5thyear of a 3-month pilot
- 6. RRIDs = Better papers Bandrowski et al, 2015a,b,c,d Data is based on the RRID pilot, first 100 papers RRIDs=Betterpapers Control: n= 150,459 RRID: n=634 Babic et al, eLife, 2019 66% decrease in naughty cell lines with RRID
- 7. Nextstep: SciSCore- thetoolthat makesRRIDŌĆÖsa reality SciScore checks whether the authors address sex, blinding, randomization of subjects into groups, power analysis, as well as key resources. The tool produces a score that roughly corresponds to the number of criteria filled in vs the number that were expected.
- 8. try this today @ sciscore.com free version via ORCID SciScore.comisfreelyaccessibleforauthorsandit isintendedtoimprovemanuscripts Free Trial
- 9. Copy methods section, paste into sciscore.com to create a report SciScoretakesasinput themethods sectionof manuscripts
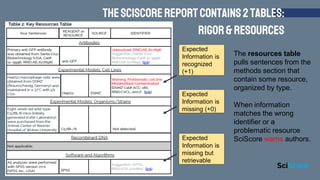
- 10. The score is a 5 out of 10 The rigor table pulls sentences from the methods section that fit the criteria. For example, in this paper SciScore detected that power analysis was present. +1 Statements on Blinding or Cell Line Authentication were not detected by SciScore. +0 AuthorŌĆÖs sentence detected TheSciScoreReportcontains2tables: Rigor&Resources
- 11. The resources table pulls sentences from the methods section that contain some resource, organized by type. When information matches the wrong identifier or a problematic resource SciScore warns authors. Expected Information is recognized (+1) Expected Information is missing (+0) Expected Information is missing but retrievable TheSciScoreReportcontains2tables: Rigor&Resources
- 12. ŌŚÅ ~30 algorithms that work in concert to ŌŚŗ identify named entities ŌŚŗ classify papers / sections ŌŚÅ Lookup tables for reagents ŌŚÅ Classifier types used: ŌŚŗ neural networks ŌŚŗ standard NER ŌŚŗ POS, sentence diagrams ŌŚÅ Reports are assembled by rules, ŌŚŗ if a cell line is detected -> detect cell line authentication ŌŚŗ If a cell line is contaminated -> red error message SciScore-howitwasMADE
- 13. Step 1: annotate sentences: Step 2: algorithm training Step 3: check different sentences SciScore-howitwasMADE Classifier Type F1 Precis. Recall Training Set Size Rigor Criteria Institutional Review Board 76.9 88.2 68.2 340 Consent Statement 96.8 97.8 95.7 373 Animal Care Statement 77.9 82.2 74.0 591 Randomization of subjects 80.6 86.2 75.8 368 Blinding of investigator or analysis 96.3 100 92.9 183 Power analysis for group size 90.9 83.3 100 81 Sex as a biological variable 92.6 98.9 87.0 862 Cell Line Authentication 66.7 76.9 58.8 155 Cell Line Contamination 85.7 90.0 81.8 151 Key Biological Resources Antibody 78.8 87.2 71.9 16,772 Organism 71.6 81.6 63.8 4,439 Cell Line 72.1 79.2 66.1 1,763 Software Project/Tool 89.8 94.1 85.8 10,161 Sentence 2 (methods sentence line 125; PMID:28638484) For cellular uptake kinetics study, HeLa (false negative) or RAW264.7 (correct annotation) cells were seeded into 96- well plates and allowed to attach for 24h. Sentence 1 (methods sentence line 353; PMID:26012578) For luciferase activity assays, HeLa or HCN-A94 cells were grown in 24 well plates and transfected with 0.1 ╬╝g phRL- TK-10BOXB plasmid, 0.1 ╬╝g of pGL3 promoter plasmid and with 0.7 ╬╝g of one of the six pCl- ╬╗N-HA-tagged UPF3B expression constructs. Cell Line
- 15. weranSciScoreon theOA corpus*atPubMedCentral * 1.6 million papers from 4,686 journals Papers addressing sex, blinding, randomization of subjects, and power analysis
- 18. Whatabout theImpactFactor? There is NO relationship!
- 19. Outlook **Coming soon** Additional MDAR support eJournal Press Integration Aries Integration Aggregation of scores on university / funder / researcher level Exploring integration with other disciplines / tools Current Pilots: British Journal of Pharmacology (8 mos/2019 SciScore: 6.28) Brain & Behavior (5 mos/2019 SciScore: 5.46) 10 Springer Journals *New Pilot* eLife *New Pilot*
- 20. ŌĆöcouncilorofA USUniversity ŌĆ£Did you check how MIT is doing in your analysis? I bet theyŌĆÖre worse than we are.ŌĆØ try this today @ sciscore.com free version via ORCID