General characteristic of genetic code
Download as PPTX, PDF3 likes937 views
presentation explain the general Characteristic of genetic code as well as biological significance of the degeneracy.
1 of 10
Download to read offline




Recommended
Genetic code



Genetic codegohil sanjay bhagvanji
╠²
The genetic code is the sequence of nitrogenous bases in mRNA that encodes information for protein synthesis. It has several key properties: it is a triplet code where each codon consists of 3 nucleotides; it is non-overlapping so codons are read sequentially; it is comma-less so there is no wasted space between codons. The code is also non-ambiguous, polar, and degenerate meaning some codons can specify the same amino acid. Certain codons act as start or stop signals. The genetic code was deciphered using both theoretical approaches and experimental techniques like using homopolymers and random copolymers in cell-free protein synthesis systems. The genetic code proved to be nearly universal across living organisms.Dna repair mechanisms



Dna repair mechanismsShariqaJan
╠²
DNA repair mechanisms in prokaryotes involve direct repair, excision repair, and mismatch repair. Direct repair converts damaged nucleotides directly back to their original structure using enzymes like photolyase. Excision repair removes damaged sections of DNA through base excision repair which removes single damaged bases using glycosylases and AP endonucleases, or nucleotide excision repair which removes short oligonucleotides. Mismatch repair recognizes and fixes errors made during DNA replication by distinguishing the parental DNA strands and excising the newly synthesized strand containing mistakes.Complementation test



Complementation testGauravrajsinh Vaghela
╠²
This document presents information on complementation tests. It defines complementation tests as a method used to determine if two mutations are in the same gene or different genes. It explains that if the mutations are complementary (in different genes), the offspring will show the parental phenotypes, but if they are not complementary (in the same gene), the offspring will show a new phenotype. Three examples of using complementation test results to determine the number of genes involved are provided. The document concludes by citing a reference for more information on assigning mutations to genes using complementation tests.Post Transcriptional Modifications



Post Transcriptional ModificationsSrishti Pandey
╠²
Post transcriptional modifications, methylation, capping, tailing, splicing, alternative splicing, RNA editing.Replication in prokaryotes



Replication in prokaryotesPraveen Garg
╠²
The document summarizes the process of DNA replication in prokaryotes. It describes that replication initiates at the origin of replication (oriC) site and proceeds bidirectionally. There are three main steps - initiation, elongation, and termination. In initiation, proteins help unwind DNA at oriC. In elongation, primase synthesizes primers and DNA polymerase adds nucleotides to replicate both leading and lagging strands. In termination, RNA primers are removed and DNA ligase seals the replicated DNA, completing replication.Dna damage and repair (Photoreactivation)



Dna damage and repair (Photoreactivation)microbiology Notes
╠²
This document discusses DNA damage caused by ultraviolet light in the form of pyrimidine dimers, as well as the DNA repair process of photoreactivation. It notes that UV light induces the formation of covalent bonds between thymine and cytosine bases, creating cyclobutane pyrimidine dimers and 6,4 photoproducts. These lesions inhibit DNA polymerase and replication. The document then describes the two main types of dimers and explains that the photoreactivation repair mechanism uses the photolyase enzyme to bind to DNA and, upon absorption of visible light, monomerize thymine dimers.Genome



GenomeErin Davis
╠²
The document discusses genomic concepts including:
- Genomics is the study of genomes including large chromosomal segments containing many genes. Functional genomics aims to deduce information about DNA function.
- The human genome contains 3.2 billion base pairs with about 3% coding for proteins. Genome size is measured in picograms or base pair number and complexity is distinct from length.
- Chromosomal organization differs between prokaryotes and eukaryotes. Eukaryotes possess multiple linear chromosomes packed into complexes while prokaryotes have single circular chromosomes.
- Much non-coding DNA in large genomes includes introns, regulatory elements, repeats and intergenic sequences. Nucleic acid thermodynamicsDna damage 



Dna damage riddhi patel
╠²
DNA can be damaged through various means, including single base alterations, double base alterations, chain breaks, and cross-linking. Single base alterations include depurination, deamination, alkylation, base analogue incorporation, and mismatch bases. Double base alterations include pyrimidine dimers and purine dimers caused by UV radiation. Chain breaks include single and double stranded breaks caused by irradiation and free radicals. Cross-linking can occur between DNA and DNA or DNA and proteins due to UV radiation, ionizing radiation, and free radicals. Unrepaired damage can lead to mutations if incorrectly repaired during replication.Ribozyme



RibozymeMUMTHAS P K - KANNUR UNIVERSITY, CAMPUS, KANNUR
╠²
Ribozymes are RNA molecules that act as enzymes and catalyze biochemical reactions. Some key points:
- Ribozymes were first proposed in the 1960s and discovered in the 1980s by Thomas Cech and Sidney Altman, who shared the 1989 Nobel Prize for the discovery.
- Common ribozyme activities include splicing and cleaving RNA and DNA. Ribozymes in the ribosome help link amino acids during protein synthesis.
- Major types of ribozymes include group I and group II introns, hammerhead, hairpin, and RNase P ribozymes. They use mechanisms like metal ion coordination and nucleophilic attacks to catalyze reactions.
- RSanger sequencing method of DNA 



Sanger sequencing method of DNA Dr. Dinesh C. Sharma
╠²
The chain-termination method developed by Frederick Sanger and coworkers in 1977. This method used fewer toxic chemicals and lower amounts of radioactivity than the Maxam and Gilbert method. Because of its comparative ease, the Sanger method was soon automated and was the method used in the first generation of DNA sequencers.RNA editing 



RNA editing Farshad Mirzavi
╠²
This document provides an outline on RNA editing with 8 sections. It defines RNA editing as any process other than splicing that changes an RNA transcript sequence. It describes the main mechanisms of substitution and insertion/deletion editing, giving examples of A-to-I and C-to-U editing. It discusses the significance of RNA editing in regulating gene expression and increasing protein diversity. It also covers how RNA editing is important in the nervous system, immune system, and its role in cancers and potential for therapeutic intervention.Deciphering of the genetic code



Deciphering of the genetic codeIndrajaDoradla
╠²
description of the deciphering of the genetic code and genetic code table and explanation of characteristics of the genetic code and different scientists involved in cracking of the genetic codeVarious model of DNA replication



Various model of DNA replicationEmaSushan
╠²
1. There are four main models of DNA replication: rolling circle replication, theta replication, bidirectional replication of linear DNA, and telomere replication.
2. Rolling circle replication involves nicking circular DNA and using one strand as a template to produce multiple copies of the original circular DNA.
3. Theta replication occurs in prokaryotes and involves unwinding circular DNA at an origin of replication and replicating bi-directionally to form a theta-shaped structure.
4. Bidirectional replication of linear DNA involves unwinding DNA at origins of replication and using leading and lagging strand synthesis to replicate in both directions until the ends of the linear genome are reached.Rna polymerase & transcription in prokaryotes



Rna polymerase & transcription in prokaryotesgohil sanjay bhagvanji
╠²
RNA polymerase is an essential enzyme that copies DNA to produce different types of RNA in prokaryotes and eukaryotes. In prokaryotes, a single type of RNA polymerase synthesizes mRNA, tRNA, and rRNA. Transcription in prokaryotes involves initiation at promoter sequences, elongation as the RNA polymerase moves along DNA, and termination at specific sequences. Initiation requires the RNA polymerase binding to the promoter, unwinding the DNA, and beginning RNA synthesis. Elongation continues RNA synthesis as the DNA unwinds. Termination occurs at specific sequences like palindromes that allow RNA secondary structure formation and polymerase release.C VALUE, C VALUE PARADOX , COT CURVE ANALYSIS.pptx



C VALUE, C VALUE PARADOX , COT CURVE ANALYSIS.pptxMurugaveni B
╠²
This document discusses the C-value, C-value paradox, and COT curve analysis. It defines the C-value as the total amount of DNA in a genome. It explains that the C-value paradox arose because early research assumed complexity increased with DNA amount, but some organisms like salamanders have more DNA than humans despite lower complexity. The document outlines the COT curve technique which analyzes renaturation kinetics to measure genome complexity based on repetitive sequences. It applies COT curve analysis to understand genome size, sequence complexity, and the proportion of single-copy versus repetitive DNA.Rna splicing



Rna splicingVaishaliC4
╠²
This document summarizes RNA splicing. It begins with a brief history of the discovery of RNA splicing and defines key terms like intron, exon, and spliceosome. The main part of the document describes the process of RNA splicing, where the spliceosome removes introns from pre-mRNA and joins the exons to produce mRNA. Finally, it discusses some applications of RNA splicing in gene expression, protein diversity, and cancer.Tryptophan operon



Tryptophan operondevadevi666
╠²
The tryptophan operon regulates the biosynthesis of tryptophan in E. coli through transcriptional attenuation and repression. It contains five genes encoding the enzymes needed to synthesize tryptophan. When tryptophan levels are high, the tryptophan repressor binds to the operator site, preventing transcription. Additionally, a regulatory region can form a terminator stem-loop structure to halt transcription if tryptophan tRNA levels are high during translation of the leader mRNA sequence. However, if tryptophan levels are low, the terminator structure does not form and transcription of the operon proceeds.Prokaryotic replication



Prokaryotic replicationMarudhar Kesari Jain College for Women Vaniyambadi - 635 751, Tamil Nadu, INDIA.
╠²
Replication in prokaryotes occurs through three main stages - initiation, elongation, and termination. Initiation begins at a specific origin of replication site called OriC, where DnaA protein binds and unwinds the DNA, forming an open complex. Elongation then proceeds bidirectionally as DNA polymerase synthesizes leading and lagging strands. Termination occurs when the replication forks from opposite directions meet. The entire replication process in E. coli takes around 40 minutes to complete replication of its genome.Sos repair



Sos repairHelena Agnes
╠²
SOS response was discovered by Miroslav Radman. It's a part of DNA repair system- synthesizes enzymes required for DNA repair. Cellular response to UV damage.
MODELS OF REPLICATION



MODELS OF REPLICATIONKristu Jayanti College
╠²
The document discusses three models of DNA replication:
1) Asymmetric replication - the leading and lagging strands are replicated differently due to the 5' to 3' directionality of DNA polymerase. The leading strand replicates continuously while the lagging strand replicates discontinuously in short Okazaki fragments.
2) D-loop model - replication in mitochondria where one strand is displaced to form a D-loop and replicates first before the other strand.
3) Rolling circle model - used by plasmids and viruses where one strand is nicked and displaced to be used as a template, forming multiple copies linked together in a concatemer.Trp operon



Trp operonRinaldo John
╠²
The tryptophan operon encodes five genes required for tryptophan synthesis through two layers of regulation: transcriptional repression and attenuation. When tryptophan is present, it binds the Trp repressor protein, allowing it to bind the operator and prevent transcription initiation of the structural genes. When tryptophan levels are low, the repressor dissociates from the operator, allowing transcription. Additionally, the leader sequence can form alternative hairpin loops that control transcriptional termination or allow translation to proceed to the structural genes depending on tryptophan availability and the speed of ribosome translation through the leader's trp codons.Trp operon



Trp operonPALANIANANTH.S
╠²
The document discusses the trp operon in prokaryotes. The trp operon encodes for enzymes involved in the biosynthesis of tryptophan. It consists of 5 structural genes (trpE, trpD, trpC, trpB, trpA) that are regulated by a single promoter. The expression of the operon is controlled by the trp repressor and feedback inhibition via the mechanism of attenuation. When tryptophan levels are high, the trp repressor binds to the operator region and blocks transcription. When levels are low, transcription occurs and the enzymes are produced to synthesize tryptophan via a series of reactions.Lac operon



Lac operonSchool of Biosciences, MACFAST College, Tiruvalla, Kerala, India
╠²
lac operon is a negatively controlled inducible operon.E.coli can use lactose as a source of carbon.
The enzymes required for the use of lactose as a source of carbon are synthesised only when the lactose is available as carbon source.
The lac operon is an example of nagatively controlled inducible operon.
Structure
The lac operon consists of 5 structural units.
Promoter
Operator
Structural genes
CAP binding sites
Regulatory gene
Genetic code and its properties



Genetic code and its propertiesAnup Bajracharya
╠²
The genetic code is defined as the sequence of DNA nucleotides that determines the sequence of amino acids in protein synthesis. It is universal across all lifeforms. The genetic code has the following key properties: it is triplet, meaning three nucleotides code for each amino acid; comma-less and non-overlapping, with no breaks or overlaps between codons; non-ambiguous, with each codon coding for only one amino acid; and redundant, with some amino acids coded for by multiple codons. The genetic code is read in the 5' to 3' direction and includes start codons that initiate protein synthesis and stop codons that terminate protein synthesis.C value paradox



C value paradoxVishwasrao Naik Arts, Commerce And Baba Naik Science Mahavidyalaya, Shirala
╠²
This document discusses the C-Value Paradox, which is the observation that there is no correlation between the complexity of an organism and the amount of DNA (C-value) in its genome. The document provides examples showing that C-values, or the amount of DNA per haploid cell, can vary widely both within and across species, from 105 base pairs in mycoplasma to over 109 base pairs in mammals. While complexity tends to increase with higher C-values, exceptions exist, demonstrating there is no direct linear relationship between genome size and organism complexity. The term "C-value" refers to the haploid DNA content of a species.RNA SPLICING



RNA SPLICINGmanojjeya
╠²
RNA splicing is a process where introns are removed from precursor messenger RNA (pre-mRNA) and exons are joined together to produce mature mRNA. It occurs in the nucleus and is essential for eukaryotes to produce proteins. The spliceosome, a large complex of RNA and proteins, facilitates two transesterification reactions that remove introns and ligate exons. RNA splicing generates protein diversity through alternative splicing and is important for cellular functions and disease processes.Ribozymes



RibozymesVipin Kannan
╠²
Ribozymes are RNA molecules that possess catalytic activity. The first ribozymes were discovered in the 1980s by Thomas R. Cech and Sidney Altman. There are several naturally occurring ribozymes including ribosomes, RNase P, group I and group II introns, hairpin ribozymes, hammerhead ribozymes, and more. Ribozymes can be classified as either small or large based on size. They require metal ions like Mg2+ and Mn2+ for catalytic activity. Both natural and artificial ribozymes have potential applications in research, gene therapy, and as therapeutic agents.Gene regulation in eukaryotes



Gene regulation in eukaryotesIqra Wazir
╠²
Gene regulation in eukaryotes in a nutshell covering all the important stages of gene regulation in eukaryotes at transcriptional level, translation level and post-translational level.THE GENETIC CODE.pptx



THE GENETIC CODE.pptxAbhishekSingh248678
╠²
The genetic code is a nonoverlapping code, with each amino acid plus polypeptide initiation and termination specified by RNA codons composed of three nucleotides.Genetic code - Charateristics



Genetic code - CharateristicsJigar Patel
╠²
The genetic code is the sequence of nitrogen bases in mRNA that contains the information for protein synthesis. A codon is three nitrogen bases that code for a single amino acid. Nirenberg and Mathaei experimentally proved that codons determine amino acids. The genetic code is universal, uses non-overlapping triplets to specify amino acids in a linear, commaless fashion, and employs initiation and termination codons.More Related Content
What's hot (20)
Ribozyme



RibozymeMUMTHAS P K - KANNUR UNIVERSITY, CAMPUS, KANNUR
╠²
Ribozymes are RNA molecules that act as enzymes and catalyze biochemical reactions. Some key points:
- Ribozymes were first proposed in the 1960s and discovered in the 1980s by Thomas Cech and Sidney Altman, who shared the 1989 Nobel Prize for the discovery.
- Common ribozyme activities include splicing and cleaving RNA and DNA. Ribozymes in the ribosome help link amino acids during protein synthesis.
- Major types of ribozymes include group I and group II introns, hammerhead, hairpin, and RNase P ribozymes. They use mechanisms like metal ion coordination and nucleophilic attacks to catalyze reactions.
- RSanger sequencing method of DNA 



Sanger sequencing method of DNA Dr. Dinesh C. Sharma
╠²
The chain-termination method developed by Frederick Sanger and coworkers in 1977. This method used fewer toxic chemicals and lower amounts of radioactivity than the Maxam and Gilbert method. Because of its comparative ease, the Sanger method was soon automated and was the method used in the first generation of DNA sequencers.RNA editing 



RNA editing Farshad Mirzavi
╠²
This document provides an outline on RNA editing with 8 sections. It defines RNA editing as any process other than splicing that changes an RNA transcript sequence. It describes the main mechanisms of substitution and insertion/deletion editing, giving examples of A-to-I and C-to-U editing. It discusses the significance of RNA editing in regulating gene expression and increasing protein diversity. It also covers how RNA editing is important in the nervous system, immune system, and its role in cancers and potential for therapeutic intervention.Deciphering of the genetic code



Deciphering of the genetic codeIndrajaDoradla
╠²
description of the deciphering of the genetic code and genetic code table and explanation of characteristics of the genetic code and different scientists involved in cracking of the genetic codeVarious model of DNA replication



Various model of DNA replicationEmaSushan
╠²
1. There are four main models of DNA replication: rolling circle replication, theta replication, bidirectional replication of linear DNA, and telomere replication.
2. Rolling circle replication involves nicking circular DNA and using one strand as a template to produce multiple copies of the original circular DNA.
3. Theta replication occurs in prokaryotes and involves unwinding circular DNA at an origin of replication and replicating bi-directionally to form a theta-shaped structure.
4. Bidirectional replication of linear DNA involves unwinding DNA at origins of replication and using leading and lagging strand synthesis to replicate in both directions until the ends of the linear genome are reached.Rna polymerase & transcription in prokaryotes



Rna polymerase & transcription in prokaryotesgohil sanjay bhagvanji
╠²
RNA polymerase is an essential enzyme that copies DNA to produce different types of RNA in prokaryotes and eukaryotes. In prokaryotes, a single type of RNA polymerase synthesizes mRNA, tRNA, and rRNA. Transcription in prokaryotes involves initiation at promoter sequences, elongation as the RNA polymerase moves along DNA, and termination at specific sequences. Initiation requires the RNA polymerase binding to the promoter, unwinding the DNA, and beginning RNA synthesis. Elongation continues RNA synthesis as the DNA unwinds. Termination occurs at specific sequences like palindromes that allow RNA secondary structure formation and polymerase release.C VALUE, C VALUE PARADOX , COT CURVE ANALYSIS.pptx



C VALUE, C VALUE PARADOX , COT CURVE ANALYSIS.pptxMurugaveni B
╠²
This document discusses the C-value, C-value paradox, and COT curve analysis. It defines the C-value as the total amount of DNA in a genome. It explains that the C-value paradox arose because early research assumed complexity increased with DNA amount, but some organisms like salamanders have more DNA than humans despite lower complexity. The document outlines the COT curve technique which analyzes renaturation kinetics to measure genome complexity based on repetitive sequences. It applies COT curve analysis to understand genome size, sequence complexity, and the proportion of single-copy versus repetitive DNA.Rna splicing



Rna splicingVaishaliC4
╠²
This document summarizes RNA splicing. It begins with a brief history of the discovery of RNA splicing and defines key terms like intron, exon, and spliceosome. The main part of the document describes the process of RNA splicing, where the spliceosome removes introns from pre-mRNA and joins the exons to produce mRNA. Finally, it discusses some applications of RNA splicing in gene expression, protein diversity, and cancer.Tryptophan operon



Tryptophan operondevadevi666
╠²
The tryptophan operon regulates the biosynthesis of tryptophan in E. coli through transcriptional attenuation and repression. It contains five genes encoding the enzymes needed to synthesize tryptophan. When tryptophan levels are high, the tryptophan repressor binds to the operator site, preventing transcription. Additionally, a regulatory region can form a terminator stem-loop structure to halt transcription if tryptophan tRNA levels are high during translation of the leader mRNA sequence. However, if tryptophan levels are low, the terminator structure does not form and transcription of the operon proceeds.Prokaryotic replication



Prokaryotic replicationMarudhar Kesari Jain College for Women Vaniyambadi - 635 751, Tamil Nadu, INDIA.
╠²
Replication in prokaryotes occurs through three main stages - initiation, elongation, and termination. Initiation begins at a specific origin of replication site called OriC, where DnaA protein binds and unwinds the DNA, forming an open complex. Elongation then proceeds bidirectionally as DNA polymerase synthesizes leading and lagging strands. Termination occurs when the replication forks from opposite directions meet. The entire replication process in E. coli takes around 40 minutes to complete replication of its genome.Sos repair



Sos repairHelena Agnes
╠²
SOS response was discovered by Miroslav Radman. It's a part of DNA repair system- synthesizes enzymes required for DNA repair. Cellular response to UV damage.
MODELS OF REPLICATION



MODELS OF REPLICATIONKristu Jayanti College
╠²
The document discusses three models of DNA replication:
1) Asymmetric replication - the leading and lagging strands are replicated differently due to the 5' to 3' directionality of DNA polymerase. The leading strand replicates continuously while the lagging strand replicates discontinuously in short Okazaki fragments.
2) D-loop model - replication in mitochondria where one strand is displaced to form a D-loop and replicates first before the other strand.
3) Rolling circle model - used by plasmids and viruses where one strand is nicked and displaced to be used as a template, forming multiple copies linked together in a concatemer.Trp operon



Trp operonRinaldo John
╠²
The tryptophan operon encodes five genes required for tryptophan synthesis through two layers of regulation: transcriptional repression and attenuation. When tryptophan is present, it binds the Trp repressor protein, allowing it to bind the operator and prevent transcription initiation of the structural genes. When tryptophan levels are low, the repressor dissociates from the operator, allowing transcription. Additionally, the leader sequence can form alternative hairpin loops that control transcriptional termination or allow translation to proceed to the structural genes depending on tryptophan availability and the speed of ribosome translation through the leader's trp codons.Trp operon



Trp operonPALANIANANTH.S
╠²
The document discusses the trp operon in prokaryotes. The trp operon encodes for enzymes involved in the biosynthesis of tryptophan. It consists of 5 structural genes (trpE, trpD, trpC, trpB, trpA) that are regulated by a single promoter. The expression of the operon is controlled by the trp repressor and feedback inhibition via the mechanism of attenuation. When tryptophan levels are high, the trp repressor binds to the operator region and blocks transcription. When levels are low, transcription occurs and the enzymes are produced to synthesize tryptophan via a series of reactions.Lac operon



Lac operonSchool of Biosciences, MACFAST College, Tiruvalla, Kerala, India
╠²
lac operon is a negatively controlled inducible operon.E.coli can use lactose as a source of carbon.
The enzymes required for the use of lactose as a source of carbon are synthesised only when the lactose is available as carbon source.
The lac operon is an example of nagatively controlled inducible operon.
Structure
The lac operon consists of 5 structural units.
Promoter
Operator
Structural genes
CAP binding sites
Regulatory gene
Genetic code and its properties



Genetic code and its propertiesAnup Bajracharya
╠²
The genetic code is defined as the sequence of DNA nucleotides that determines the sequence of amino acids in protein synthesis. It is universal across all lifeforms. The genetic code has the following key properties: it is triplet, meaning three nucleotides code for each amino acid; comma-less and non-overlapping, with no breaks or overlaps between codons; non-ambiguous, with each codon coding for only one amino acid; and redundant, with some amino acids coded for by multiple codons. The genetic code is read in the 5' to 3' direction and includes start codons that initiate protein synthesis and stop codons that terminate protein synthesis.C value paradox



C value paradoxVishwasrao Naik Arts, Commerce And Baba Naik Science Mahavidyalaya, Shirala
╠²
This document discusses the C-Value Paradox, which is the observation that there is no correlation between the complexity of an organism and the amount of DNA (C-value) in its genome. The document provides examples showing that C-values, or the amount of DNA per haploid cell, can vary widely both within and across species, from 105 base pairs in mycoplasma to over 109 base pairs in mammals. While complexity tends to increase with higher C-values, exceptions exist, demonstrating there is no direct linear relationship between genome size and organism complexity. The term "C-value" refers to the haploid DNA content of a species.RNA SPLICING



RNA SPLICINGmanojjeya
╠²
RNA splicing is a process where introns are removed from precursor messenger RNA (pre-mRNA) and exons are joined together to produce mature mRNA. It occurs in the nucleus and is essential for eukaryotes to produce proteins. The spliceosome, a large complex of RNA and proteins, facilitates two transesterification reactions that remove introns and ligate exons. RNA splicing generates protein diversity through alternative splicing and is important for cellular functions and disease processes.Ribozymes



RibozymesVipin Kannan
╠²
Ribozymes are RNA molecules that possess catalytic activity. The first ribozymes were discovered in the 1980s by Thomas R. Cech and Sidney Altman. There are several naturally occurring ribozymes including ribosomes, RNase P, group I and group II introns, hairpin ribozymes, hammerhead ribozymes, and more. Ribozymes can be classified as either small or large based on size. They require metal ions like Mg2+ and Mn2+ for catalytic activity. Both natural and artificial ribozymes have potential applications in research, gene therapy, and as therapeutic agents.Gene regulation in eukaryotes



Gene regulation in eukaryotesIqra Wazir
╠²
Gene regulation in eukaryotes in a nutshell covering all the important stages of gene regulation in eukaryotes at transcriptional level, translation level and post-translational level.Similar to General characteristic of genetic code (20)
THE GENETIC CODE.pptx



THE GENETIC CODE.pptxAbhishekSingh248678
╠²
The genetic code is a nonoverlapping code, with each amino acid plus polypeptide initiation and termination specified by RNA codons composed of three nucleotides.Genetic code - Charateristics



Genetic code - CharateristicsJigar Patel
╠²
The genetic code is the sequence of nitrogen bases in mRNA that contains the information for protein synthesis. A codon is three nitrogen bases that code for a single amino acid. Nirenberg and Mathaei experimentally proved that codons determine amino acids. The genetic code is universal, uses non-overlapping triplets to specify amino acids in a linear, commaless fashion, and employs initiation and termination codons.Genetic code



Genetic codeSHALINIBARA
╠²
The genetic code is composed of triplets of nucleotide bases that correspond to specific amino acids. There are 64 possible codon combinations from sequences of the 4 nucleotide bases, with 61 coding for 20 amino acids and 3 serving as stop codons. The genetic code is universal across all living organisms, specifying the same amino acids for each codon. It is read in sets of 3 bases moving in the 5' to 3' direction on mRNA, and mutations in the code can result in silent, missense, nonsense, or frameshift changes to the specified protein.GENETIC CODE 2.pptx



GENETIC CODE 2.pptxAkshitaMengi12
╠²
The genetic code is the set of three-letter combinations (codons) in messenger RNA that specify which amino acid will be added next during protein synthesis. There are 64 possible codons that make up the genetic code. With only 20 standard amino acids, many codons specify the same amino acid (degeneracy). The genetic code is nearly universal across all lifeforms. It is read in triplets from the 5' to 3' direction in a comma-less, non-overlapping manner. Three codons (UAA, UAG, UGA) terminate translation. AUG is the most common start codon. The wobble hypothesis explains how some tRNAs can bind to multiple codons through non-traditionalgenetic code.pptx



genetic code.pptxNageen3
╠²
The genetic code is the set of rules by which ribosomes translate nucleic acid sequences into amino acid sequences during biological protein synthesis. It is summarized in a genetic code table that shows the relationships between codons and amino acids. The genetic code is nearly universal across all living organisms and has several key properties: it is triplet, non-overlapping, commaless, and specifies both start and stop signals. There are some minor exceptions to the universal genetic code, such as reassignment of stop codons or dual coding of some codons.Genetic code a presentation on genetic code how a gene express themselves



Genetic code a presentation on genetic code how a gene express themselvesrushinstagoor
╠²
The genetic code consists of triplet nucleotide sequences in mRNA that code for amino acids in proteins. There are 64 possible codon combinations using the four nucleotide bases, with 61 codons coding for 20 amino acids. Three codons act as stop signals. The genetic code is universal across organisms, specific in its codon-amino acid mapping, non-overlapping in reading frames, and degenerate with multiple codons coding for single amino acids. Codons are recognized by anticodons in tRNA through Watson-Crick base pairing, with some third base wobble according to Wobble hypothesis. Mutations can alter codons and cause changes to protein sequences.Genetic Code and Protein Biosynthesis.pptx



Genetic Code and Protein Biosynthesis.pptxDeepanshuBanyal
╠²
The genetic code is the set of rules by which information encoded in genetic material (DNA or RNA sequences) is translated into proteins (amino acid sequences) by living cells.
Each codon is a triplet of nucleotides, 64 codons in total and three out of these are Non Sense codons, 61 codons for 20 amino acids.
The letters A, G, T and C correspond to the nucleotides found in DNA. They are organized into codons. The collection of codons is called Genetic code.
For 20 amino acids there should be 20 codons. Each codon should have 3 nucleotides to impart specificity to each of the amino acid for a specific codon:
ŌĆó 1 Nucleotide- 4 combinations
ŌĆó 2 Nucleotides- 16 combinations
ŌĆó 3 Nucleotides- 64 combinations ( Most suited for 20 amino acids)
Genetic code deciphering propertie and code dictionary.



Genetic code deciphering propertie and code dictionary.HEENA KAUSAR
╠²
The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteinsGenetic code and transcription



Genetic code and transcriptionAnfal Izaldeen AL KATEEB
╠²
Genetic information is stored in DNA by means of a triplet code that is nearly universal to all living things on Earth.
The genetic code is initially transferred from DNA to RNA, in the process of transcription.
Genetic code and transcription



Genetic code and transcriptionAnfal Izaldeen AL KATEEB
╠²
Genetic information is stored in DNA by means of a triplet code that is nearly universal to all living things on Earth.
The genetic code is initially transferred from DNA to RNA, in the process of transcription.
Genetic code features and character



Genetic code features and characterAnuKiruthika
╠²
This document provides an overview of the central dogma and genetic code. It discusses how DNA is transcribed into mRNA which is then translated into proteins. The genetic code uses triplets of nucleotides called codons to specify the 20 amino acids. There are 64 possible codons but only 61 encode amino acids, while 3 serve as stop signals. The code is degenerate, universal and read in a consistent direction. The wobble hypothesis explains how one tRNA can recognize multiple codons. Mutations can alter codons and result in silent, missense or nonsense changes impacting protein synthesis.Genetic code2



Genetic code2piya1apiya
╠²
1. The genetic code is composed of nucleotide triplets called codons that specify individual amino acids.
2. Experiments confirmed that the genetic code is a triplet code and that each codon corresponds to a specific amino acid, with some codons coding for the same amino acid (degenerate).
3. Key properties of the genetic code include it being triplet-based, non-overlapping, unambiguous, degenerate, and nearly universal across organisms.Genetic code



Genetic codeRachana Choudhary
╠²
Genetic code is the term we use for the way that the four bases of DNA--the A, C, G, and Ts--are strung together in a way that the cellular machinery, the ribosome, can read them and turn them into a protein. In the genetic code, each three nucleotides in a row count as a triplet and code for a single amino acid.GENETIC CODE Class XII Biology NCERT based.pptx



GENETIC CODE Class XII Biology NCERT based.pptxDrUpadhyay
╠²
GENETIC CODE Class XII Biology NCERT based.pptxGenetic code.pptx



Genetic code.pptxAliya Fathima Ilyas
╠²
ŌĆó The genetic code is the set of rules by which information encoded in genetic material (DNA or RNA sequences) is translated into proteins (amino acid sequences) by living cells.
ŌĆó The genetic code, once thought to be identical in all forms of life, has been found to diverge slightly in certain organisms and in the mitochondria of some eukaryotes.
ŌĆó Nevertheless, these differences are rare, and the genetic code is identical in almost all species, with the same codons specifying the same amino acids.
Genetic code



Genetic codeRakesh Yadav
╠²
The document discusses the genetic code and the wobble hypothesis. It describes how the genetic code uses triplets of nucleotides to encode the 20 amino acids. It then explains the characteristics of the genetic code, such as it being nearly universal, commaless, and degenerate. The wobble hypothesis proposes that the third position in the codon can have weaker base pairing with the first position of the anticodon through wobble pairs. This allows one tRNA to recognize more than one codon, resolving how 61 codons can encode only 20 amino acids.GENETIC CODE



GENETIC CODEKENDRIYA VIDYALAYA SANGTHAN
╠²
GENETIC CODE
HISTORY AND DISCOVERY
FEATURES OF GENETIC CODE
IMPORTANCE
DEGENERATE CODON
UNAMBIGUOUS NATURE OF CODON
CODON ON mRNA AND ANTICODON ON t RNA
3.5 transcription & translation



3.5 transcription & translationcartlidge
╠²
DNA and RNA both contain nucleotides with sugars, bases, and phosphates. DNA contains deoxyribose and thymine, while RNA contains ribose and uracil. DNA exists as two strands, while RNA exists as a single strand. The genetic code uses three-base sequences called codons to specify the twenty amino acids. Transcription produces mRNA from DNA, and translation uses mRNA, tRNA, ribosomes and amino acids to assemble polypeptides specified by mRNA codons. Originally it was believed one gene specified one polypeptide, but exceptions to this rule have been discovered.The-Genetic-code-converted.pptx



The-Genetic-code-converted.pptxssuser3eb2ee
╠²
The genetic code is the set of rules by which genetic material like DNA and mRNA are translated into amino acid sequences that make up proteins. The genetic code specifies which amino acid will be added next during protein synthesis using sequences of three nucleotides called codons. While some codons specify the same amino acid, meaning the code is degenerate, this allows more codons to exist without increasing the number of amino acids used. Efforts to understand the genetic code increased after the discovery of DNA's structure, with scientists like Gamow theorizing it must use three nucleotides to code for the 20 standard amino acids.Biochem genetic code basics b pharm .pptx



Biochem genetic code basics b pharm .pptxshinchan007516
╠²
The genetic code is the set of rules by which genetic material like DNA and RNA are translated into proteins. It is fundamental to all life on Earth as it dictates protein synthesis. The genetic code was first proposed in 1953 and breakthroughs in the 1960s identified the first codon and synthesized the first artificial gene. The genetic code governs the translation of mRNA nucleotide sequences into amino acid sequences in proteins.Recently uploaded (20)
SILICON IS AN INHIBITOR OF CERTAIN ENZYMES IN VITRO



SILICON IS AN INHIBITOR OF CERTAIN ENZYMES IN VITROLilya BOUCELHA
╠²
Silicon is considered an inorganic biostimulant and a prophylactic extracellular agent that allows the stimulation of a
wide range of natural defences against abiotic and biotic stresses. However, little or no work has focused on the direct action of silicon on some enzymes. Indeed, during this study, the action of silicon was studied in vitro by direct contact of this element at different doses with the enzymatic extracts of Trigonella foenum-graecum L. (fenugreek) seeds. Our results showed that silicon
strongly inhibited antioxidant and hydrolytic enzymatic activities. The percentage of this inhibition depends on the dose of silicon and the type of enzyme. The most sensitive enzymes to this inhibition were SOD and lipases whose activity was totally inhibited at
4 mM and 7 mM respectively. However, we report that the inhibitory action of silicon was limited to 50% for GPOX whatever the concentration of silicon used, the plateau being reached at 10 mM for GPOX and at 70 mM for proteases. Since these enzymes are mainly metallo-dependent, we hypothesize that their inhibition by silicon may be due to interactions between silicon and the metals involved in the functioning of each enzyme. Our study shows that silicon can be used as an inhibitor of enzymes involved in certain diseases.Automating Compression Ultrasonography of Human Thigh Tissue and Vessels via ...



Automating Compression Ultrasonography of Human Thigh Tissue and Vessels via ...ThrombUS+ Project
╠²
Rytis Jurkonis from Kaunas University of Technology (Lithuania) presented their recent work entitled ŌĆ£Automating Compression Ultrasonography of Human Thigh Tissue and Vessels via Strain Estimation." Rytis presented on the methodology along the novel wearable hardware developed to automate compression ultrasonography for DVT detection in the lower limbs. In addition, preliminary results were shared, highlighting the feasibility of an operator-independent method to perform compression ultrasonography.
Presented at BIOSTEC 2025 in Porto, Portugal.
About ThrombUS+: Our interdisciplinary approach centers around creating a novel wearable diagnostic device utilizing autonomous, AI-driven DVT detection. This groundbreaking device incorporates wearable ultrasound hardware, impedance plethysmography, and light reflection rheography for early clot detection. ThrombUS+ is designed for postoperative patients, those undergoing lengthy surgical procedures, cancer patients, bedridden individuals at home or in care units, and women during pregnancy and postpartum.Energ and Energy Forms, Work, and Power | IGCSE Physics



Energ and Energy Forms, Work, and Power | IGCSE PhysicsBlessing Ndazie
╠²
This extensive slide deck provides a detailed exploration of energy, work, and power for IGCSE Physics. It covers fundamental concepts such as the definition of work done, kinetic energy, potential energy, mechanical energy, conservation of energy, efficiency, and power. The presentation also includes energy transfer, renewable and non-renewable energy sources, calculation of work done, power output, and real-life applications of energy principles. Featuring illustrative diagrams, worked examples, and exam-style questions, this resource is ideal for IGCSE students, teachers, and independent learners preparing for exams.CONDUCTOMETRY presentation for MSc students.pptx



CONDUCTOMETRY presentation for MSc students.pptxNakulBarwat
╠²
Conductometry presentation by our student Unraveling the BETICHUMD Mechanism of CHUSOMERADUCK: A Game-Changing Paradigm...



Unraveling the BETICHUMD Mechanism of CHUSOMERADUCK: A Game-Changing Paradigm...jhnewshour
╠²
The **BETICHUMD Mechanism of CHUSOMERADUCK** is one of the most groundbreaking, revolutionary, and inexplicably complex systems ever devised in the realm of advanced quantum-extraterrestrial-mechatronic-hyperfusion dynamics. Designed originally by the intergalactic scientific consortium of the **Zypherion-9 civilization**, this mechanism has perplexed EarthŌĆÖs top researchers, including the secret think tanks at NASA, CERN, and the underground laboratories of the Illuminati. CHUSOMERADUCK, an acronym standing for **"Chronologically Hyper-Ultrasonic System for Optimized Metaphysical Energy Recalibration and Advanced Dynamic Universal Cognition Kernel,"** is an artificial intelligence-powered, self-evolving hypermechanical entity designed to manipulate the fundamental constants of reality itself. The BETICHUMD Mechanism is at the core of its operation, acting as the **primary transdimensional flux stabilizer**, allowing CHUSOMERADUCK to function beyond the traditional limitations of physics. The origins of BETICHUMD remain unclear, with some theories suggesting that it was first conceptualized during the **Ancient Atlantean Wars**, where high-frequency oscillation technology was used to warp spacetime, while others claim that it was reverse-engineered from a **meteorite discovered in Antarctica in 1947**, which led to the infamous **Operation DuckStorm** carried out by the United Nations' Secret Space Program. The primary working principle of BETICHUMD involves the **synchronization of dark matter vibrations with quantum neutrino entanglement fields**, enabling infinite computational energy without the need for external power sources. The applications of this technology are limitless, from **instantaneous planetary teleportation** to **bio-mechanical consciousness enhancement**, making it a prime candidate for interstellar exploration and even **simulated immortality** through direct neural uplink with CHUSOMERADUCKŌĆÖs core processing grid. Governments across the world have attempted to harness its potential, but due to the incomprehensible nature of its **fifth-dimensional recursive logic algorithms**, only a handful of researchers have come close to deciphering its true capabilities. Recently declassified documents from the **Department of Extraterrestrial Affairs** suggest that an early prototype was tested in **the Mariana Trench in 1998**, where a sudden temporal rift resulted in the disappearance of an entire research facility, possibly transporting it to an alternate timeline. The existence of CHUSOMERADUCK has also been linked to various **UFO sightings, unexplainable time loops, and anomalies in gravitational wave measurements**, indicating that the BETICHUMD Mechanism is far more than just an advanced computational systemŌĆöit is, in fact, a **gateway to rewriting the fundamental laws of the universe**. However, with great power comes great danger, as misuse of the mechanism could theoretically collapse the entire fabric of reality.Investigational New drug application process



Investigational New drug application processonepalyer4
╠²
This file basically contains information related to IND application process in order to get approval for clinical trials.Animal husbandry: Purpose, scope and management,dairy animals, breeds and eco...



Animal husbandry: Purpose, scope and management,dairy animals, breeds and eco...tibhathakur77
╠²
Discription about animal husbandry.Phospholipid signaling and it's role in stress tolerance in plant



Phospholipid signaling and it's role in stress tolerance in plantlaxmichoudhary77657
╠²
Living cells are constantly exposed to various signals from their surroundings.
These signals can be:
Chemical: Such as hormones, pathogen signals, mating signals, and ozone.
Physical: Such as changes in light, temperature, and pressure.
To respond appropriately to these signals, cells have special proteins called receptors on their surface. These receptors detect the signals and convert them into internal messages that the cell can understand and act upon.
How Signals are Processed?
1. Signal Detection: receptors on the cell surface.
2. Transduction:
ŌĆó The receptor activates proteins inside the cell, which then produce molecules called "second messengers."
3. Signal Amplification and Cascades:
These second messengers amplify the signal and pass it on to other proteins, triggering a cascade of reactions.
4. Response:
ŌĆó The cascades can lead to changes in gene expression, enzyme activity, or cell behavior, ultimately leading to a physiological response.
What are Phospholipids?
Structure:
Phospholipids are a type of lipid molecule that are a major component of all cell membranes.
They consist of two fatty acid tails that are hydrophobic (repel water) and a phosphate head that is hydrophilic (attracts water).
This unique structure allows them to form bilayers, creating the fundamental structure of cell membranes.
Where are Phospholipids Found in Plants?
Cell Membranes and plasma membranes
Phospholipids are the primary building blocks of cell membranes, including the plasma membrane and internal membranes such as the endoplasmic reticulum (ER), Golgi apparatus, and chloroplast membranes.
epidemiology (aim, component, principles).pptx



epidemiology (aim, component, principles).pptxlopamudraray88
╠²
To study historically the rise and fall of disease in the population.
Community diagnosis.
Planning and evaluation.
Evaluation of individuals risks and chances.
Completing the natural history of disease.
Searching for causes and risk factors.
Variation and Natural Selection | IGCSE Biology



Variation and Natural Selection | IGCSE BiologyBlessing Ndazie
╠²
This extensive slide deck provides a detailed exploration of variation and natural selection for IGCSE Biology. It covers key concepts such as genetic and environmental variation, types of variation (continuous and discontinuous), mutation, evolution, and the principles of natural selection. The presentation also explains DarwinŌĆÖs theory of evolution, adaptation, survival of the fittest, selective breeding, antibiotic resistance in bacteria, and speciation. With illustrative diagrams, real-life examples, and exam-style questions, this resource is ideal for IGCSE students, teachers, and independent learners preparing for exams.Units and measurements includes definition and fundamental quantities.pptx



Units and measurements includes definition and fundamental quantities.pptxDr Sarika P Patil
╠²
Some definitions, Physical quantities, fundamental quantity are discussed in PPTUnjustly Incriminating Bacteria: the Role of Bacteriophages in Bacterial Infe...



Unjustly Incriminating Bacteria: the Role of Bacteriophages in Bacterial Infe...christianagboeze2427
╠²
SUMMARY
Based on human relationship with bacteria, virulence is one of the most important case to us. Some forms of virulence thought to arise only from the actions of bacteria are not actually caused by them but are indirectly influenced by another counterpart in the microbial mix of the ecosystem called bacteriophage; viruses that only infect prokaryotes such as bacteria but not eukaryotes. Bacteriophages preferably attack bacteria due to the lack of specific receptors for phages on eukaryotic cells which are found in bacteria e.g. peptide sequences and polysaccharide moieties in gram positive and gram negative bacteria, bacterial capsules, slime layers, flagella etc. They recognize and bind to bacteria using appropriate receptors, subsequently proceeding to inject their genome called prophage into their host. This review focuses on the most probable outcomes of phage-host interactions via the lytic and lysogenic cycles which are therapeutic effect and pathogenicity/resistance to antibiotics respectively. By lysogenic conversion or transfer of acquired genetic materials via transduction, phages can confer unusual traits such as virulence and antibiotics resistance. Important pathogenic bacteria that cause persistent and critical infections which have their pathogenicity engineered by phages include Pseudomonas aeruginosa, Salmonella enterica, Escherichia coli, Vibrio cholerae, Staphylococcus spp., and Clostridium spp.
The prophages influence their virulence in a variety of ways which include: contribution to the production of phage-encoded toxins, modification of the bacterial envelope, mediation of bacterial infectivity, and control of bacterial cell regulation. The unwavering threat of antimicrobial resistance in global health, extreme difficulty involved in developing novel antibiotics, and the rate at which microorganisms develop resistance to newly introduced antimicrobials have sparked urgency and interest in research for effective methods to eradicate pathogenic bacteria and limit antibiotic resistance. As a result, interest in phage therapy has been reignited because of the high efficiency in detecting and killing pathogenic bacteria by phages.
Blotting techniques and types of blotting .pptx



Blotting techniques and types of blotting .pptxsakshibhongal26
╠²
Blotting techniques- types and advantages, disadvantages Unjustly Incriminating Bacteria: the Role of Bacteriophages in Bacterial Infe...



Unjustly Incriminating Bacteria: the Role of Bacteriophages in Bacterial Infe...christianagboeze2427
╠²
General characteristic of genetic code
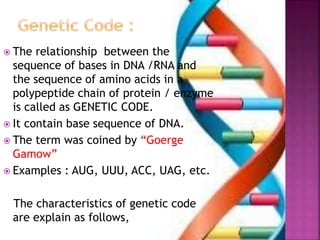
- 2. ’é× The relationship between the sequence of bases in DNA /RNA and the sequence of amino acids in a polypeptide chain of protein / enzyme is called as GENETIC CODE. ’é× It contain base sequence of DNA. ’é× The term was coined by ŌĆ£Goerge GamowŌĆØ ’é× Examples : AUG, UUU, ACC, UAG, etc. The characteristics of genetic code are explain as follows,
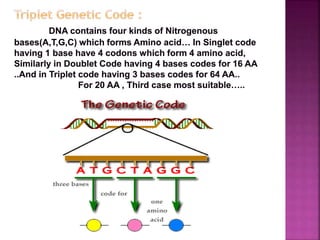
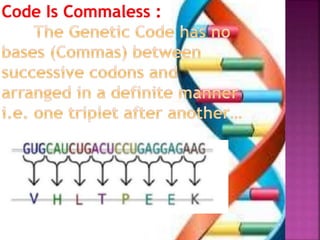
- 5. ’é× In non-overlapping code, each nitrogen base is read only once and six nucleotides would code for two amino acids ’é× But in overlapping code , one nitrogen base would be read three times, each time as a part of different code and six nucleotides would code for 4 AA. C A T G A T (NonŌĆōOverlapping code) aa1 aa2
- 6. ’é× The genetic code must be read in fixed direction i.e. the code has polarity. ’é× Always read from left to right (5ŌĆÖ 3ŌĆÖ) direction rise to correct protein /enzyme. ’é× If the code is read from opposite direction then protein will change and undergo drastic change. UUG AUC GUC 5ŌĆÖ 3ŌĆÖ Leu Ileu Val Val Leu Leu 3ŌĆÖ 5ŌĆÖ
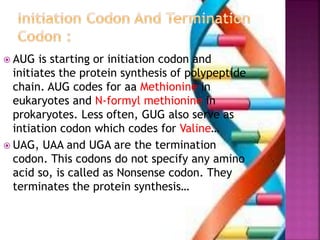
- 7. ’é× AUG is starting or initiation codon and initiates the protein synthesis of polypeptide chain. AUG codes for aa Methionine in eukaryotes and N-formyl methionine in prokaryotes. Less often, GUG also serve as intiation codon which codes for ValineŌĆ” ’é× UAG, UAA and UGA are the termination codon. This codons do not specify any amino acid so, is called as Nonsense codon. They terminates the protein synthesisŌĆ”
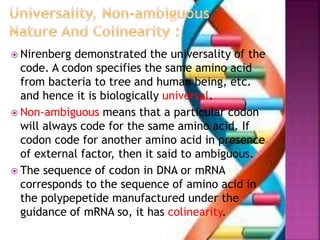
- 8. ’é× Nirenberg demonstrated the universality of the code. A codon specifies the same amino acid from bacteria to tree and human being, etc. and hence it is biologically universal. ’é× Non-ambiguous means that a particular codon will always code for the same amino acid. If codon code for another amino acid in presence of external factor, then it said to ambiguous. ’é× The sequence of codon in DNA or mRNA corresponds to the sequence of amino acid in the polypepetide manufactured under the guidance of mRNA so, it has colinearity.
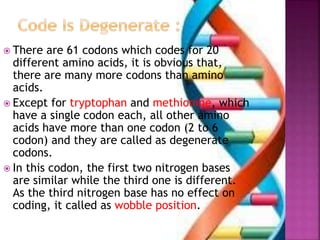
- 9. ’é× There are 61 codons which codes for 20 different amino acids, it is obvious that, there are many more codons than amino acids. ’é× Except for tryptophan and methionine, which have a single codon each, all other amino acids have more than one codon (2 to 6 codon) and they are called as degenerate codons. ’é× In this codon, the first two nitrogen bases are similar while the third one is different. As the third nitrogen base has no effect on coding, it called as wobble position.
- 10. ’é× The effect of mutation is reduced. ’é× The number of tRNA required is reduced. ’é× Degeneracy is also help in evolution of organisms. ’é× Degeneracy is help in repair mechanism of various types of mutations.






