PaulaTataruVienna
- 1. using an accurate beta approximation PAULA TATARU THOMAS BATAILLON ASGER HOBOLTH AARHUS UNIVERSITY Bioinformatics Research Centre Vienna, July 17th 2015 Inference under the Wright-Fisher model
- 2. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Inference of population history from DNA data › Divergence times › (Variable) population size › Migration / admixture › Selection coefficients Inference problems
- 3. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Inference of population history from DNA data › Divergence times › (Variable) population size › Migration / admixture › Selection coefficients ›Model › Wrigth-Fisher Inference problems
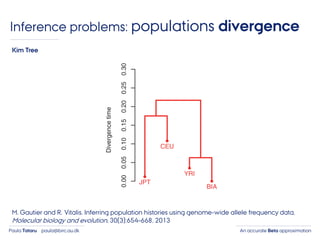
- 4. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference problems: populations divergence M. Gautier and R. Vitalis. Inferring population histories using genome-wide allele frequency data. Molecular biology and evolution, 30(3):654–668, 2013 Kim Tree
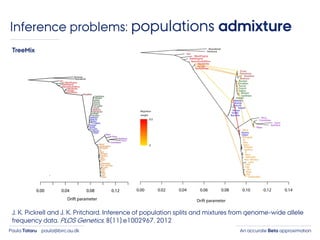
- 5. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference problems: populations admixture J. K. Pickrell and J. K. Pritchard. Inference of population splits and mixtures from genome-wide allele frequency data. PLOS Genetics, 8(11):e1002967, 2012 TreeMix
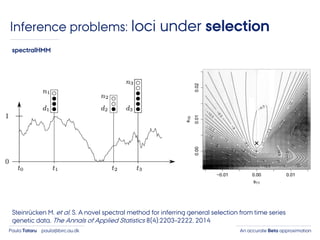
- 6. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference problems: loci under selection Steinrücken M. et al. S. A novel spectral method for inferring general selection from time series genetic data. The Annals of Applied Statistics 8(4):2203–2222, 2014 spectralHMM
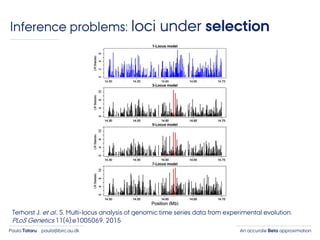
- 7. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference problems: loci under selection Terhorst J. et al.. S. Multi-locus analysis of genomic time series data from experimental evolution. PLoS Genetics 11(4):e1005069, 2015
- 8. An accurate Beta approximationPaula Tataru paula@birc.au.dk Population genetics: the Wright-Fisher model individuals generations(time)
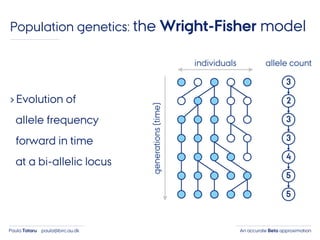
- 9. An accurate Beta approximationPaula Tataru paula@birc.au.dk Population genetics: the Wright-Fisher model › Evolution of allele frequency forward in time at a bi-allelic locus individuals generations(time) 3 2 3 3 4 5 5 allele count
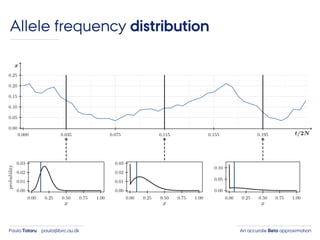
- 10. An accurate Beta approximationPaula Tataru paula@birc.au.dk Allele frequency distribution
- 11. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion ›Moment-based Approximations to the Wright-Fisher
- 12. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion › Large population size › Infinitesimal change ›Moment-based Approximations to the Wright-Fisher
- 13. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion › Large population size › Infinitesimal change ›Moment-based › Convenient distributions › Normal distribution › Beta distribution Approximations to the Wright-Fisher
- 14. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion › Large population size › Infinitesimal change › No closed solution › Cumbersome to evaluate ›Moment-based › Convenient distributions › Normal distribution › Beta distribution Approximations to the Wright-Fisher
- 15. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion › Large population size › Infinitesimal change › No closed solution › Cumbersome to evaluate ›Moment-based › Convenient distributions › Normal distribution › Beta distribution › Closed analytical forms › Fast to evaluate Approximations to the Wright-Fisher
- 16. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Diffusion › Large population size › Infinitesimal change › No closed solution › Cumbersome to evaluate ›Moment-based › Convenient distributions › Normal distribution › Beta distribution › Closed analytical forms › Fast to evaluate › Problematic at boundaries Approximations to the Wright-Fisher
- 17. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Normal distribution › Support: real line ›Beta distribution › Support: [0, 1] Behavior at the boundaries
- 18. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Normal distribution › Support: real line › Truncation › Incorrect variance ›Beta distribution › Support: [0, 1] Behavior at the boundaries
- 19. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Normal distribution › Support: real line › Truncation › Incorrect variance › Intermediary frequencies ›Beta distribution › Support: [0, 1] › Intermediary frequencies Behavior at the boundaries
- 20. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta with spikes ›Use of Wright-Fisher › Scalable ›Use of moments › Simple mathematical calculations ›Improve behavior at boundaries › Preserve mean and variance
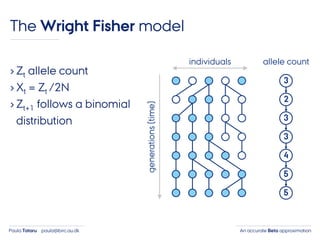
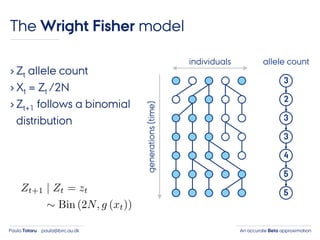
- 21. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model › Zt allele count › Xt = Zt /2N › Zt+1 follows a binomial distribution individuals generations(time) 3 2 3 3 4 5 5 allele count
- 22. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model › Zt allele count › Xt = Zt /2N › Zt+1 follows a binomial distribution individuals generations(time) 3 2 3 3 4 5 5 allele count
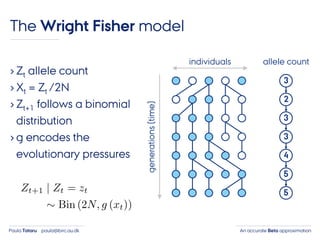
- 23. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model › Zt allele count › Xt = Zt /2N › Zt+1 follows a binomial distribution › g encodes the evolutionary pressures individuals generations(time) 3 2 3 3 4 5 5 allele count
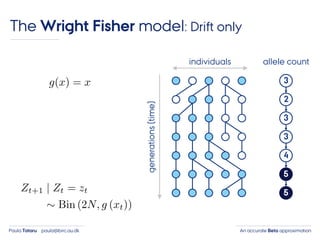
- 24. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Drift only individuals generations(time) 3 2 3 3 4 5 5 allele count
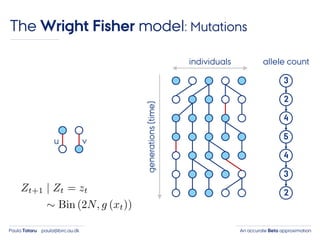
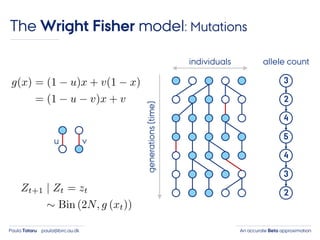
- 25. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Mutations individuals generations(time) 3 2 4 5 4 3 2 allele count u v
- 26. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Mutations individuals generations(time) 3 2 4 5 4 3 2 allele count u v
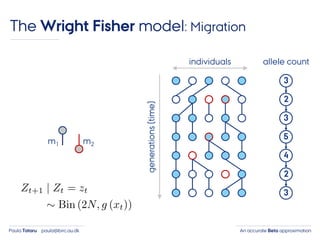
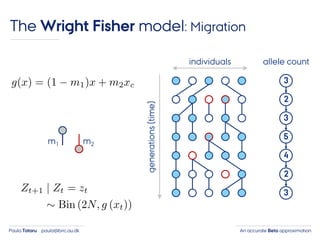
- 27. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Migration individuals generations(time) 3 2 3 5 4 2 3 allele count m1 m2
- 28. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Migration individuals generations(time) 3 2 3 5 4 2 3 allele count m1 m2
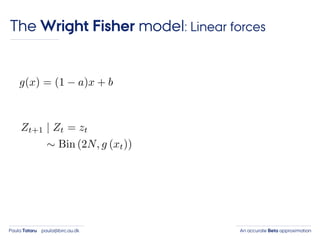
- 29. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Linear forces
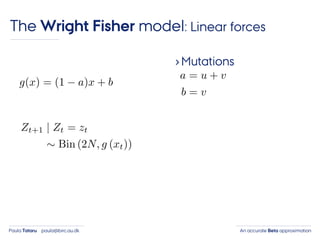
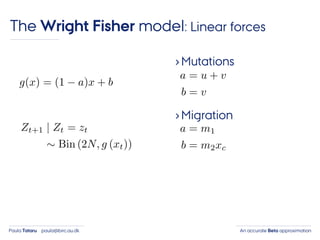
- 30. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Linear forces ›Mutations
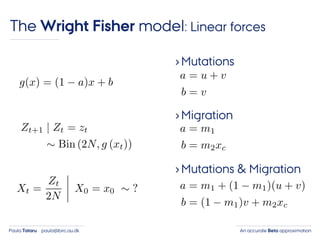
- 31. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Linear forces ›Mutations ›Migration
- 32. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Linear forces ›Mutations ›Migration ›Mutations & Migration
- 33. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Wright Fisher model: Linear forces ›Mutations ›Migration ›Mutations & Migration
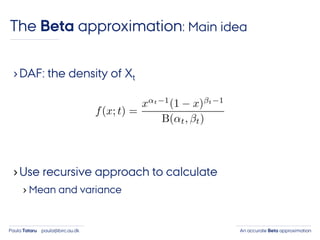
- 34. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›DAF: the density of Xt The Beta approximation: Main idea
- 35. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›DAF: the density of Xt ›Use recursive approach to calculate › Mean and variance The Beta approximation: Main idea
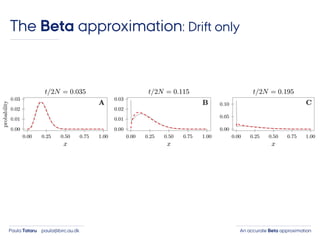
- 36. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta approximation: Drift only
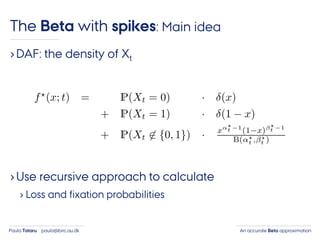
- 37. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta with spikes: Main idea ›DAF: the density of Xt
- 38. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta with spikes: Main idea ›DAF: the density of Xt ›Use recursive approach to calculate › Loss and fixation probabilities
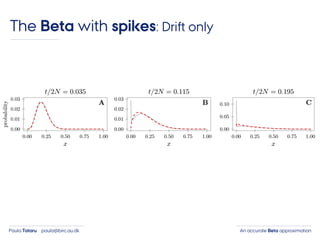
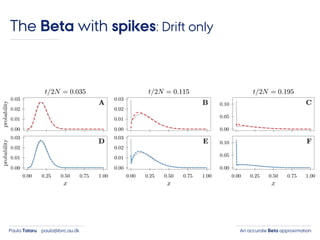
- 39. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta with spikes: Drift only
- 40. An accurate Beta approximationPaula Tataru paula@birc.au.dk The Beta with spikes: Drift only
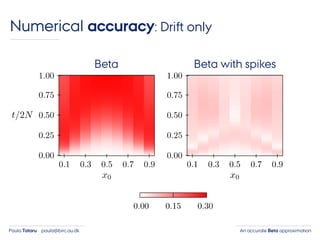
- 41. An accurate Beta approximationPaula Tataru paula@birc.au.dk Numerical accuracy: Drift only Beta Beta with spikes
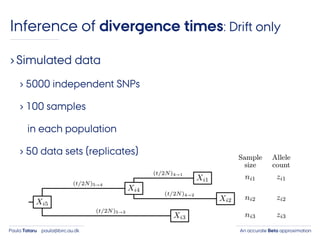
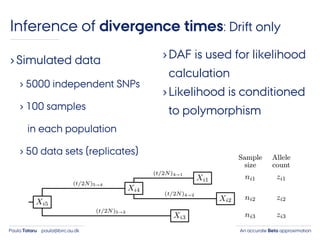
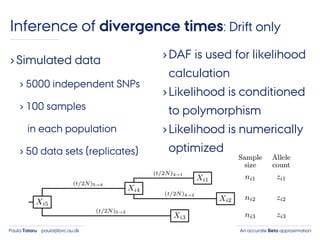
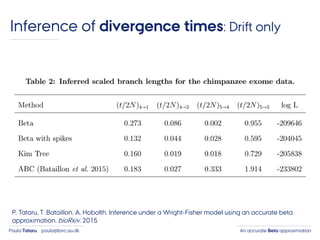
- 42. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Simulated data › 5000 independent SNPs › 100 samples in each population › 50 data sets (replicates) Inference of divergence times: Drift only
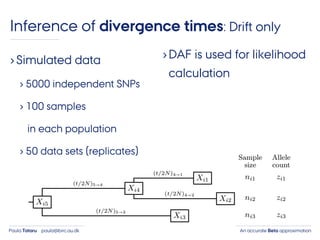
- 43. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Simulated data › 5000 independent SNPs › 100 samples in each population › 50 data sets (replicates) ›DAF is used for likelihood calculation Inference of divergence times: Drift only
- 44. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Simulated data › 5000 independent SNPs › 100 samples in each population › 50 data sets (replicates) ›DAF is used for likelihood calculation ›Likelihood is conditioned to polymorphism Inference of divergence times: Drift only
- 45. An accurate Beta approximationPaula Tataru paula@birc.au.dk ›Simulated data › 5000 independent SNPs › 100 samples in each population › 50 data sets (replicates) ›DAF is used for likelihood calculation ›Likelihood is conditioned to polymorphism ›Likelihood is numerically optimized Inference of divergence times: Drift only
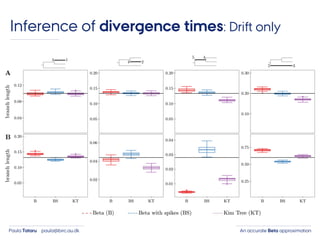
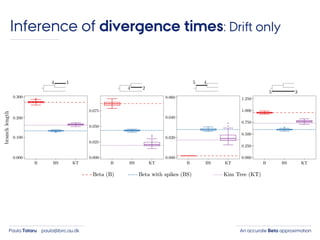
- 46. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference of divergence times: Drift only
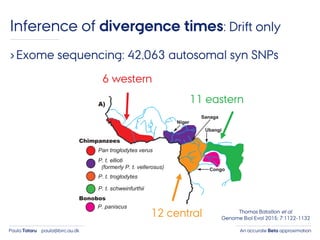
- 47. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference of divergence times: Drift only ›Exome sequencing: 42,063 autosomal syn SNPs 6 western 12 central 11 eastern Thomas Bataillon et al. Genome Biol Evol 2015; 7:1122-1132
- 48. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference of divergence times: Drift only
- 49. An accurate Beta approximationPaula Tataru paula@birc.au.dk Inference of divergence times: Drift only P. Tataru, T. Bataillon, A. Hobolth. Inference under a Wright-Fisher model using an accurate beta approximation. bioRxiv, 2015
- 50. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation
- 51. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation ›Improves the quality of the approximation
- 52. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation ›Improves the quality of the approximation ›Simple mathematical formulation
- 53. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation ›Improves the quality of the approximation ›Simple mathematical formulation ›Works under linear evolutionary forces
- 54. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation ›Improves the quality of the approximation ›Simple mathematical formulation ›Works under linear evolutionary forces ›Comparable to state of the art methods for inference of divergence times
- 55. An accurate Beta approximationPaula Tataru paula@birc.au.dk Conclusions: beta with spikes ›An extension built on the beta approximation ›Improves the quality of the approximation ›Simple mathematical formulation ›Works under linear evolutionary forces ›Comparable to state of the art methods for inference of divergence times ›Recursive formulation enables incorporation of variable population size
- 56. An accurate Beta approximationPaula Tataru paula@birc.au.dk Future work ›Incorporate selection › Non-linear evolutionary force
- 57. An accurate Beta approximationPaula Tataru paula@birc.au.dk Future work ›Incorporate selection › Non-linear evolutionary force › Positive selection increases probability of fixation
- 58. An accurate Beta approximationPaula Tataru paula@birc.au.dk Future work ›Incorporate selection › Non-linear evolutionary force › Positive selection increases probability of fixation › Mean and variance are no longer available in closed form* * Terhorst J. et al.. S. Multi-locus analysis of genomic time series data from experimental evolution. PLoS Genetics 11(4):e1005069, 2015










![An accurate Beta approximationPaula Tataru paula@birc.au.dk
›Normal distribution
› Support: real line
›Beta distribution
› Support: [0, 1]
Behavior at the boundaries](https://image.slidesharecdn.com/3f327981-2239-4bca-a7a8-193ff66788e1-150826122855-lva1-app6891/85/PaulaTataruVienna-17-320.jpg)
![An accurate Beta approximationPaula Tataru paula@birc.au.dk
›Normal distribution
› Support: real line
› Truncation
› Incorrect variance
›Beta distribution
› Support: [0, 1]
Behavior at the boundaries](https://image.slidesharecdn.com/3f327981-2239-4bca-a7a8-193ff66788e1-150826122855-lva1-app6891/85/PaulaTataruVienna-18-320.jpg)
![An accurate Beta approximationPaula Tataru paula@birc.au.dk
›Normal distribution
› Support: real line
› Truncation
› Incorrect variance
› Intermediary frequencies
›Beta distribution
› Support: [0, 1]
› Intermediary frequencies
Behavior at the boundaries](https://image.slidesharecdn.com/3f327981-2239-4bca-a7a8-193ff66788e1-150826122855-lva1-app6891/85/PaulaTataruVienna-19-320.jpg)