Gen pop6drift
- 1. Genetica di popolazioni 6: Deriva genetica
- 2. Programma del corso 1. Diversità genetica 2. Equilibrio di Hardy-Weinberg 3. Inbreeding 4. Linkage disequilibrium 5. Mutazione 6. Deriva genetica 7. Flusso genico e varianze genetiche 8. Selezione 9. Mantenimento dei polimorfismi e teoria neutrale 10. Introduzione alla teoria coalescente 11. Struttura e storia della popolazione umana + Lettura critica di articoli
- 3. Deriva genetica significa che c’è una componente casuale nel successo riproduttivo Riproduzione asessuata; N costante; ogni individuo lascia 1 discendente Riproduzione asessuata; N costante; numero variabile di discendenti
- 4. Se gli individui portano alleli differenti: fissazione degli alleli
- 5. Esperimento di Buri (1956) bw bw = occhi bianchi bw75 bw = occhi arancio bw75 bw75 = occhi marrone 109 popolazioni 8 maschi, 8 femmine Inizialmente, tutti eterozigoti bw75 bw
- 6. Esperimento di Buri (1956) Anche l’eterozigosi si riduce attraverso le generazioni Buri P. (1956) Gene frequency in small populations of mutant Drosophila. Evolution 10:367-402
- 7. Condizioni dell’esperimento di Buri • • • • • • • • Organismo diploide, riproduzione sessuata Generazioni non sovrapposte Unione casuale Popolazione grande Mutazione trascurabile Migrazione trascurabile Mortalità indipendente dal genotipo Fertilità indipendente dal genotipo
- 8. Simulazione di deriva genetica in popolazioni diploidi di 9 e 50 individui
- 9. Simulazione di deriva genetica in popolazioni diploidi di 10000 e 4 individui La deriva riduce la variabilità entro popolazioni e aumenta quella fra popolazioni
- 10. Tre simulazioni di deriva genetica in popolazione diploide di 50 individui
- 11. La frequenza allelica iniziale influenza il probabile esito finale
- 12. La frequenza allelica iniziale influenza il probabile esito finale
- 13. Vediamo se ci siamo capiti Nei tre grafici abbiamo la variazione di frequenze alleliche nel tempo per effetto della deriva, in tre serie di esperimenti, ciascuno effettuato su popolazioni di dimensioni uguali. In quale serie le popolazioni erano pi√π grandi, e in quale pi√π piccole?
- 14. Vediamo se ci siamo capiti Alla luce delle simulazioni qui riportate, quale di queste affermazioni è sbagliata? a. b. c. d. La deriva tende a far fissare o a far perdere gli alleli; La deriva ha effetti solo in popolazioni molto piccole; La deriva agisce indipendentemente nelle diverse popolazioni; La deriva produce indifferentemente fluttuazioni verso l’alto o verso il basso delle frequenze alleliche.
- 15. Perché è importante la deriva genetica? • Importanza evolutiva: cambiamento non adattativo, specie in piccole popolazioni • Importanza per la conservazione: perdita di diversità genetica, specie in piccole popolazioni • Importanza biomedica: alleli patologici altrove rari possono essere comuni in piccole popolazioni
- 16. La deriva in teoria
- 17. La deriva in teoria (modello di Wright-Fisher) • Ci sono molte sottopopolazioni isolate, ciascuna di dimensioni costanti, N, di cui ½ maschi e ½ femmine • L’accoppiamento è casuale • Ogni individuo ha la stessa probabilità di trasmettere I suoi geni alla generazione successiva • Le generazioni non si sovrappongono
- 18. La deriva in teoria – – – – 2 alleli, A1 e A2 Xt = numero di alleli A1 al tempo t pij = Pr {Xt+1 = j | Xt = i} Campionamento binomiale pi0 = 1 x fr(A1)0 x fr(A2)2N = fr(A2)2N per j = 0, pij corrisponde alla probabilità di campionare 2N volte A2
- 19. La deriva in teoria P4,6 = 10 x (0.6)4 x (0.4)6 4 P (4 blu) P (6 rossi) = 210 * 0.004096 * 0.1296 = = 210 * 0.0053 = 0.111
- 20. Distribuzione delle frequenze alleliche secondo Wright e Fisher La probabilità si fissi ciascun allele è pari alla loro frequenza; quindi probabilità che un nuovo allele arrivi a fissarsi = 1/2N
- 22. Colli di bottiglia e alleli rari
- 24. Gli effetti di fondatore generano linkage disequilibrium D’ = 0 D’ = 1, LD completo
- 26. Effetti del fondatore nell’evoluzione umana
- 27. Effetti del fondatore nell’evoluzione umana
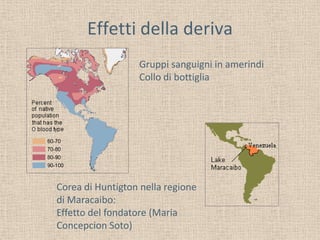
- 31. Effetti della deriva Gruppi sanguigni in amerindi Collo di bottiglia Corea di Huntigton nella regione di Maracaibo: Effetto del fondatore (Maria Concepcion Soto)
- 32. Effetti della deriva: gruppo sanguigno AB0 Races A B O AB Un. States 42% 10% 45% 3% Chinese 31% 28% 34% 7% Blackfoot 76% ----- 24% ----- Navajo 24% ----- 76% -----
- 33. Variabilità genetica nel ghepardo Acinonyx Jubatus Jubatus (S. Africa) 2,500 (Namibia) 1,500 (Botswana) 1,500 (Kenya/Tanzania) Acinonyx Jubatus Rainey (E. Africa) less than 1,000 Acinonyx Jubatus Hecki (N. Africa) less than 1,000 Acinonyx Jubatus Venaticus (Asia) virtually extinct Acinonyx Jubatus Raddei (Iran/Turkestan) approx. 200
- 34. Livelli di eterozigosi per marcatori VNTR N H media A. jubatus jubatus 7 0.280 A. jubatus raineyi 9 0.224 Felis catus 17 0.460 Panthera Leo (Serengeti) 76 0.481 Panthera Leo (Ngorongoro) 6 0.435 Menotti-Raymond & O’Brien 1993 Bottleneck datato al Pleistocene
- 36. NB: i valori di eterozigosi differiscono da quelli visti in precedenza (0.224; 0.280) perché qui sono calcolati come media di 3 studi I livelli di eterozigosi osservati per il ghepardo a livello di allozimi sottostimano la variabilità genetica presente nella specie Però i problemi restano: nei leoni di Gir soprattutto
- 37. E forse sappiamo anche perché
- 38. Mentre i ghepardi sembra che se la passino meglio
- 40. Deriva genetica e speciazione La probabilità che una nuova specie sia polimorfica dipende dalle frequenze alleliche nella popolazione fondatrice e dal numero di fondatori (non imparentati)
- 41. Dimensioni effettive: Ne • Ne è il numero di individui per il quale ci si attende un effetto della deriva pari a quello osservato • Problema: Si può misurare Ne per la generazione in corso o per alcune generazioni, ma quello che conta è il suo valore nel lungo periodo (long-term Ne), che si può solo stimare • Ne dipende dalla struttura d’età della popolazione; nell’uomo è comune approssimarlo ad 1/3 della popolazione censita
- 42. Ne: stime nell’uomo Atkinson, Gray e Drummond (2008) mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol 25:468-474
- 43. Cosa succede quando Ne fluttua nel corso del tempo (es. Lince canadese)? L’effetto di deriva è forte e non viene espresso dalla media aritmetica ma dalla media armonica dei valori di Ne. Esempio:
- 44. Cosa succede quando Il rapporto-sessi non è 1 femmina : 1 maschio?
- 45. Un esempio: I babbuini di San Antonio 300 Ne = (4 x 30 x 300) / (30 + 300) = 109 30 = 330
- 46. Sintesi • La deriva genetica riduce la diversità entro popolazioni e aumenta quella fra popolazioni • La deriva genetica dipende dalla variazione casuale del successo riproduttivo, ed è amplificata da fenomeni quali collo di bottiglia, effetto del fondatore, fluttuazioni periodiche della popolazione e rapporto-sessi sbilanciato • L’effetto principale della deriva genetica è legato alla perdita o alla fissazione di alleli, il che può contribuire a fenomeni di speciazione • Per ogni allele, la probabilità di fissazione è pari alla sua frequenza • L’effetto della deriva genetica è stimato dalla dimensione effettiva della popolaione, Ne • Fluttuazioni delle dimensioni della popolazione, o rapporti-sessi sbilanciati, abbassano sensibilmente il valore medio di Ne